S5: DOT - Image Reconstruction
[1]:
# This cells setups the environment when executed in Google Colab.
try:
import google.colab
!curl -s https://raw.githubusercontent.com/ibs-lab/cedalion/dev/scripts/colab_setup.py -o colab_setup.py
# Select branch with --branch "branch name" (default is "dev")
%run colab_setup.py
except ImportError:
pass
Notebook configuration
Decide for an example with a sparse probe or a high density probe for DOT. The notebook will load example data accordingly.
Also specify, if precomputed results of the photon propagation should be used and if the 3D visualizations should be interactive.
[2]:
# the head model used in this example
HEAD_MODEL = "colin27"
# the dataset used in this example
DATASET = "fingertappingDOT"
# used precomputed forward model results
PRECOMPUTED_SENSITIVITY = True
FORWARD_MODEL = "MCX"
# set this flag to True to enable interactive 3D plots
INTERACTIVE_PLOTS = False
[3]:
import pyvista as pv
if INTERACTIVE_PLOTS:
pv.set_jupyter_backend('server')
else:
pv.set_jupyter_backend('static')
from pathlib import Path
from tempfile import TemporaryDirectory
import cedalion
import cedalion.data
import cedalion.dataclasses as cdc
import cedalion.dot
import cedalion.dot as dot
import cedalion.io
import cedalion.sigproc.motion as motion
import cedalion.sigproc.physio as physio
import cedalion.sigproc.quality as quality
import cedalion.vis.anatomy.sensitivity_matrix
import cedalion.vis.blocks as vbx
import matplotlib.pyplot as p
import numpy as np
import xarray as xr
from cedalion import units
from cedalion.io.forward_model import load_Adot
from IPython.display import Image
xr.set_options(display_expand_data=False);
[4]:
# helper function to display gifs in rendered notbooks
def display_image(fname : str):
display(Image(data=open(fname,'rb').read(), format='png'))
Working Directory
In this notebook tHe output of the fluence and sensitivity calculations are stored in a temporary directory. This will be deleted when the notebook ends.
[5]:
temporary_directory = TemporaryDirectory()
working_directory = Path(temporary_directory.name)
Load a DOT finger-tapping dataset
loading via
cedalion.datagive stimulus events descriptive string labels
[6]:
rec = cedalion.data.get_fingertappingDOT()
rec.stim.cd.rename_events(
{
"1": "Control",
"2": "FTapping/Left",
"3": "FTapping/Right",
"4": "BallSqueezing/Left",
"5": "BallSqueezing/Right",
}
)
The location of the probes is obtained from the snirf metadata.
The units (‘mm’) are adopted and the coordinate system is named ‘digitized’.
[7]:
display(rec.geo3d)
print(f"CRS='{rec.geo3d.points.crs}'")
<xarray.DataArray (label: 346, digitized: 3)> Size: 8kB
[mm] -77.82 15.68 23.17 -61.91 21.23 56.49 ... 14.23 -38.28 81.95 -0.678 -37.03
Coordinates:
type (label) object 3kB PointType.SOURCE ... PointType.LANDMARK
* label (label) <U6 8kB 'S1' 'S2' 'S3' 'S4' ... 'FFT10h' 'FT10h' 'FTT10h'
Dimensions without coordinates: digitized
CRS='digitized'
[8]:
cedalion.vis.anatomy.plot_montage3D(rec["amp"], rec.geo3d)
Preprocessing
[9]:
rec["od"] = cedalion.nirs.cw.int2od(rec["amp"])
rec["od_tddr"] = motion.tddr(rec["od"])
rec["od_wavelet"] = motion.wavelet(rec["od_tddr"])
# see 2_tutorial_preprocessing.ipynb for channel selection
bad_channels = ['S13D26', 'S14D28']
rec["od_clean"] = rec["od_wavelet"].sel(channel=~rec["od"].channel.isin(['S13D26', 'S14D28']))
od_var = quality.measurement_variance(rec["od_clean"], calc_covariance=False)
rec["od_mean_subtracted"], global_comp = physio.global_component_subtract(
rec["od_clean"], ts_weights=1 / od_var, k=0
)
rec["od_freqfiltered"] = rec["od_mean_subtracted"].cd.freq_filter(
fmin=0.01, fmax=0.5, butter_order=4
)
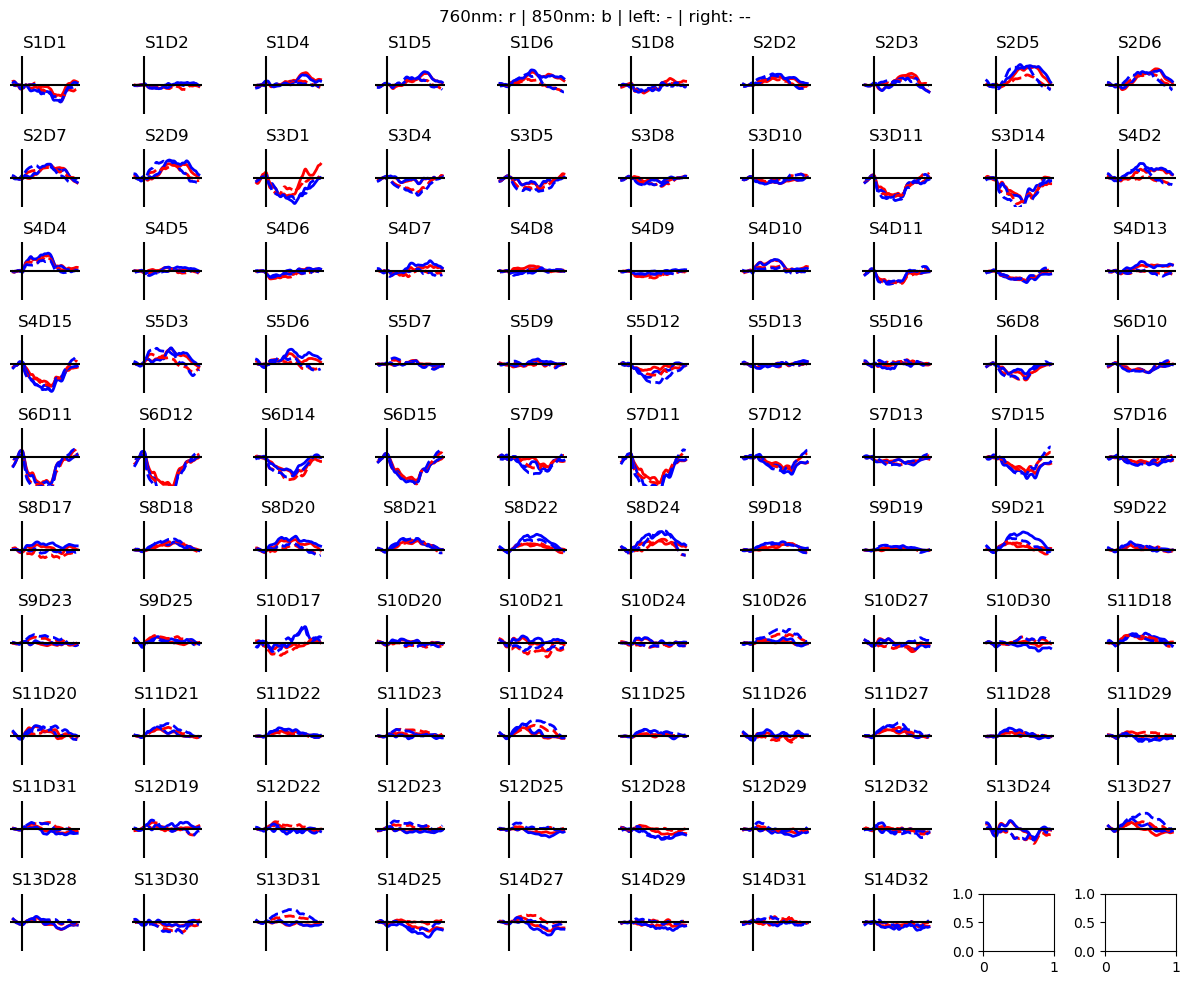
Calculate block averages in optical density
[10]:
# segment data into epochs
epochs = rec["od_freqfiltered"].cd.to_epochs(
rec.stim, # stimulus dataframe
["FTapping/Left", "FTapping/Right"], # select events, discard the others
before=5 * cedalion.units.s, # seconds before stimulus
after=30 * cedalion.units.s, # seconds after stimulus
)
# calculate baseline
baseline = epochs.sel(reltime=(epochs.reltime < 0)).mean("reltime")
# subtract baseline
epochs_blcorrected = epochs - baseline
# group trials by trial_type. For each group individually average the epoch dimension
blockaverage = epochs_blcorrected.groupby("trial_type").mean("epoch")
# Plot block averages. Please ignore errors if the plot is too small in the HD case
noPlts2 = int(np.ceil(np.sqrt(len(blockaverage.channel))))
f,ax = p.subplots(noPlts2,noPlts2, figsize=(12,10))
ax = ax.flatten()
for i_ch, ch in enumerate(blockaverage.channel):
for ls, trial_type in zip(["-", "--"], blockaverage.trial_type):
ax[i_ch].plot(blockaverage.reltime, blockaverage.sel(wavelength=760, trial_type=trial_type, channel=ch), "r", lw=2, ls=ls)
ax[i_ch].plot(blockaverage.reltime, blockaverage.sel(wavelength=850, trial_type=trial_type, channel=ch), "b", lw=2, ls=ls)
ax[i_ch].grid(1)
ax[i_ch].set_title(ch.values)
ax[i_ch].set_ylim(-.01, .01)
ax[i_ch].set_axis_off()
ax[i_ch].axhline(0, c="k")
ax[i_ch].axvline(0, c="k")
p.suptitle("760nm: r | 850nm: b | left: - | right: --")
p.tight_layout()
Constructing the TwoSurfaceHeadModel
either factory or from segmentation masks
notes about from_segmentation: The segmentation masks are in individual niftii files. The dict mask_files maps mask filenames relative to SEG_DATADIR to short labels. These labels describe the tissue type of the mask.
In principle the user is free to choose these labels. However, they are later used to lookup the tissue’s optical properties. So they must be map to one of the tabulated tissue types (c.f. cedalion.imagereco.tissue_properties.TISSUE_LABELS).
The variable landmarks_file holds the path to a file containing landmark positions in scanner space (RAS). This file can be created with Slicer3D.
[11]:
if HEAD_MODEL in ["colin27", "icbm152"]:
# use a factory method for the standard head models
head_ijk = cedalion.dot.get_standard_headmodel(HEAD_MODEL)
elif HEAD_MODEL == "custom":
# point these to a directory with segmentation masks
segm_datadir = Path("/path/to/dir/with/segmentation_masks")
mask_files = {
"csf": "mask_csf.nii",
"gm": "mask_gray.nii",
"scalp": "mask_skin.nii",
"skull": "mask_bone.nii",
"wm": "mask_white.nii",
}
# the landmarks must be in scanner (RAS) space.
# For example Slicer3D can be used to pick the landmarks.
landmarks_file = Path("/path/to/landmarks.mrk.json")
# if available provide a mapping between vertices and labels
parcel_file = None
# Construct a head model from segmentation mask.
head_ijk = cedalion.dot.TwoSurfaceHeadModel.from_segmentation(
segmentation_dir=segm_datadir,
mask_files=mask_files,
landmarks_ras_file=landmarks_file,
parcel_file=parcel_file,
# adjust these to control mesh parameters
brain_face_count=None,
scalp_face_count=None,
smoothing=0.0,
)
# Likely, better brain and scalp surfaces are achievable from
# specialized segmentation tools.
head_ijk = cedalion.dot.TwoSurfaceHeadModel.from_surfaces(
segmentation_dir=segm_datadir,
mask_files=mask_files,
landmarks_ras_file=landmarks_file,
parcel_file=parcel_file,
brain_surface_file=segm_datadir / "mask_brain.obj",
scalp_surface_file=segm_datadir / "mask_scalp.obj",
)
else:
raise ValueError("unknown head model")
Optode Registration
The optode coordinates from the recording must be aligned with the scalp surface. Currently, cedaĺion offers a simple registration method, which finds an affine transformation (scaling, rotating, translating) that matches the landmark positions of the head model and their digitized counter parts. Afterwards, optodes are snapped to the nearest vertex on the scalp.
[12]:
geo3d_snapped_ijk = head_ijk.align_and_snap_to_scalp(rec.geo3d)
display(geo3d_snapped_ijk)
<xarray.DataArray (label: 346, ijk: 3)> Size: 8kB
[] 15.1 140.9 100.2 30.75 144.6 130.6 ... 172.5 137.6 42.28 172.7 126.0 43.07
Coordinates:
type (label) object 3kB PointType.SOURCE ... PointType.LANDMARK
* label (label) <U6 8kB 'S1' 'S2' 'S3' 'S4' ... 'FFT10h' 'FT10h' 'FTT10h'
Dimensions without coordinates: ijk[13]:
plt = pv.Plotter()
vbx.plot_surface(plt, head_ijk.brain, color="w")
vbx.plot_surface(plt, head_ijk.scalp, opacity=.1)
vbx.plot_labeled_points(plt, geo3d_snapped_ijk)
plt.show()

[14]:
# remove landmarks from geo3d for subsequent plots
geo3d_plot = geo3d_snapped_ijk[geo3d_snapped_ijk.type != cdc.PointType.LANDMARK]
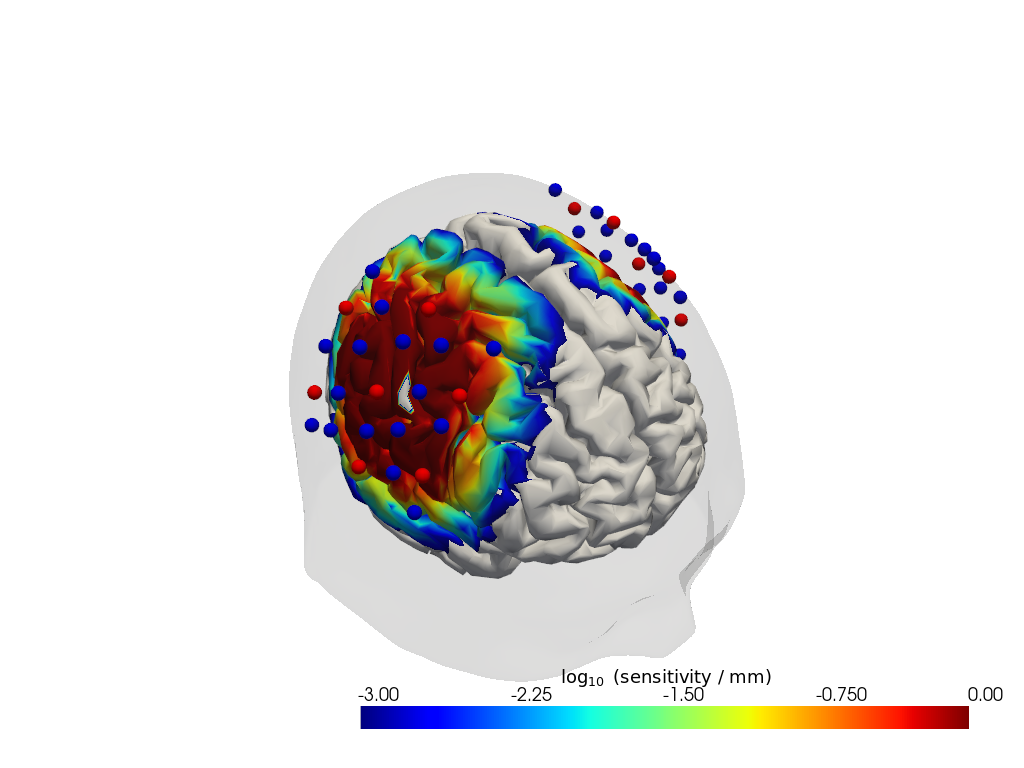
Sensitivity matrix
[15]:
Adot = cedalion.data.get_precomputed_sensitivity(DATASET, HEAD_MODEL)
Plot Sensitivity Matrix
[16]:
plotter = cedalion.vis.anatomy.sensitivity_matrix.Main(
sensitivity=Adot,
brain_surface=head_ijk.brain,
head_surface=head_ijk.scalp,
labeled_points=geo3d_plot,
)
plotter.plot(high_th=0, low_th=-3)
plotter.plt.show()
Run the image reconstruction
The class cedalion.dot.ImageRecon implements several methods to solve the inverse problem of reconstructing activations in brain and scalp tissue that would explain the measured optical density changes in channel space.
[17]:
recon = cedalion.dot.ImageRecon(
Adot,
recon_mode="mua2conc",
brain_only=False,
alpha_meas=0.01,
alpha_spatial=None,
apply_c_meas=False,
spatial_basis_functions=None,
)
In this example the channel-space block averages should be reconstructed into image space:
[18]:
blockaverage
[18]:
<xarray.DataArray (trial_type: 2, channel: 98, wavelength: 2, reltime: 154)> Size: 483kB
0.001354 0.001405 0.001391 0.001314 ... 0.0003461 0.0004625 0.0005884 0.0007154
Coordinates:
* reltime (reltime) float64 1kB -5.038 -4.809 -4.58 ... 29.54 29.77 30.0
* channel (channel) object 784B 'S1D1' 'S1D2' 'S1D4' ... 'S14D31' 'S14D32'
source (channel) object 784B 'S1' 'S1' 'S1' 'S1' ... 'S14' 'S14' 'S14'
detector (channel) object 784B 'D1' 'D2' 'D4' 'D5' ... 'D29' 'D31' 'D32'
* wavelength (wavelength) float64 16B 760.0 850.0
* trial_type (trial_type) object 16B 'FTapping/Left' 'FTapping/Right'Run the reconstruction and display results. Note how the OD time series with dimensions ('channel', 'wavelength', ...) transformed into a concentration time series with dimensions ('chromo', 'vertex', ...). The other dimensions ('trial_type', 'reltime') did not change.
The 'vertex' dimension has two coordinates:
is_brain: a boolean mask that discriminates between vertices on the brain and scalp surfacesparcel: a string label attributed to each vertex according to a brain parcellation scheme (see section ‘Parcel Space’)
[19]:
img = recon.reconstruct(blockaverage)
img
[19]:
<xarray.DataArray (chromo: 2, vertex: 25052, trial_type: 2, reltime: 154)> Size: 123MB
[µM] -2.2e-11 -1.964e-11 -1.724e-11 ... -1.489e-12 -1.522e-12 -1.547e-12
Coordinates:
* chromo (chromo) <U3 24B 'HbO' 'HbR'
parcel (vertex) object 200kB 'VisCent_ExStr_8_LH' ... 'scalp'
is_brain (vertex) bool 25kB True True True True ... False False False
* reltime (reltime) float64 1kB -5.038 -4.809 -4.58 ... 29.54 29.77 30.0
* trial_type (trial_type) object 16B 'FTapping/Left' 'FTapping/Right'
Dimensions without coordinates: vertexVisualizing image reconstruction results
In the following, different cedalion plot functions will be showcased to visualize concentration changes on the brain and scalp surfaces.
Channel Space
The function cedalion.vis.anatomy.scalp_plot_gif allows to create an animated gif of channel-space OD changes projected on the scalp.
Here, it is used to show the time course of the blockaveraged OD changes.
[20]:
# configure the plot
data_ts = blockaverage.sel(wavelength=850, trial_type="FTapping/Right")
# scalp_plot_gif expects the time dimension to be named 'time'
data_ts = data_ts.rename({"reltime": "time"})
filename_scalp = "scalp_plot_ts"
# call plot function
cedalion.vis.anatomy.scalp_plot_gif(
data_ts,
rec.geo3d,
filename=filename_scalp,
time_range=(-5, 30, 0.5) * units.s,
scl=(-0.01, 0.01),
fps=6,
optode_size=6,
optode_labels=True,
str_title="OD 850 nm",
)
[21]:
display_image(f"{filename_scalp}.gif")
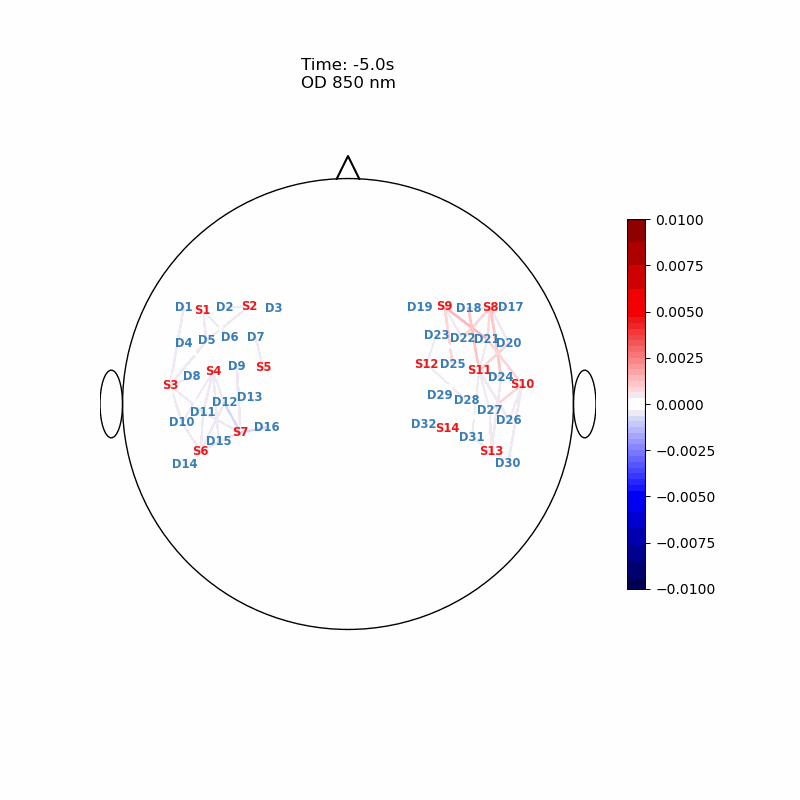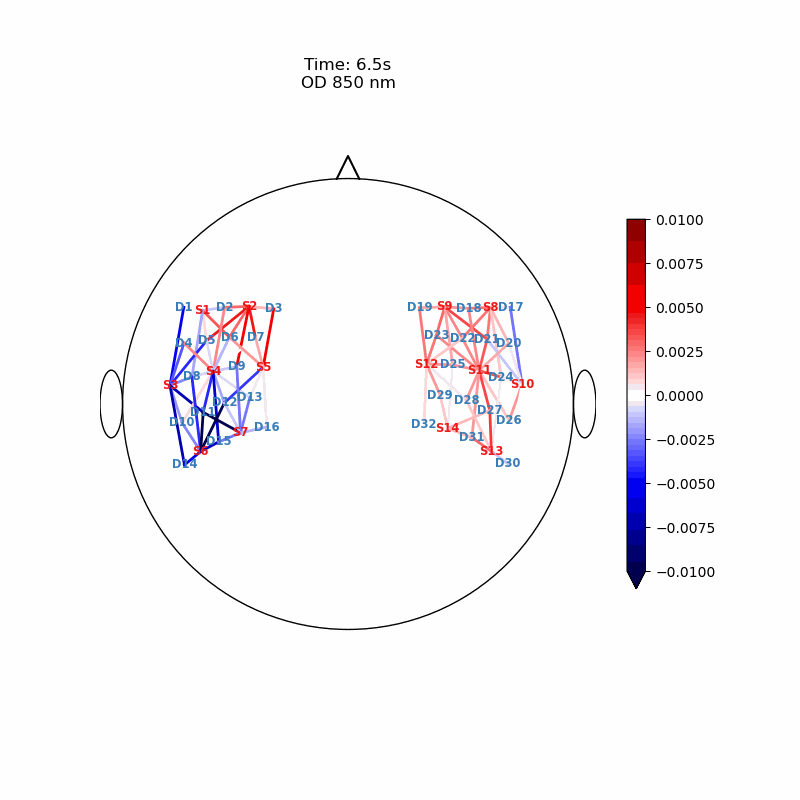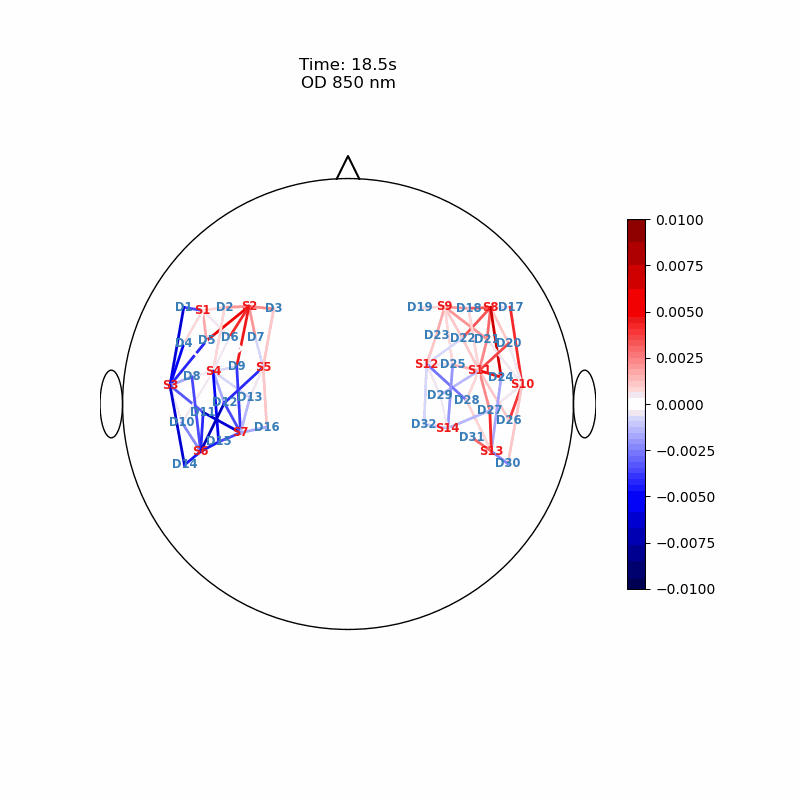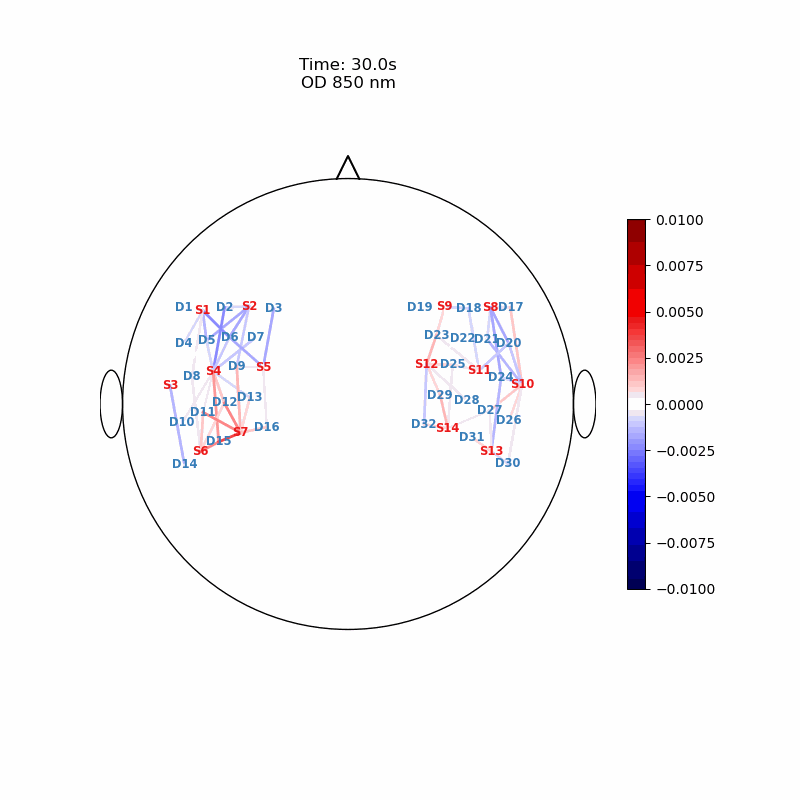
Image Space
Single-View Animations of Activitations on the Brain
The function cedalion.vis.anatomy.image_recon_view allows to create an animated gif of image-space concentration changes projected on the brain.
[22]:
filename_view = 'image_recon_view'
# image_recon_view expects input with dimensions
# ("vertex", "chromo", "time") (in that order)
X_ts = img.sel(trial_type="FTapping/Right").rename({"reltime": "time"})
X_ts = X_ts.transpose("vertex", "chromo", "time")
scl = np.percentile(np.abs(X_ts.sel(chromo='HbO')).pint.dequantify(), 99)
clim = (-scl,scl)
cedalion.vis.anatomy.image_recon_view(
X_ts, # time series data; can be 2D (static) or 3D (dynamic)
head_ijk,
cmap='seismic',
clim=clim,
view_type='hbo_brain',
view_position='left',
title_str='HbO / µM',
filename=filename_view,
SAVE=True,
time_range=(-5,30,0.5)*units.s,
fps=6,
geo3d_plot = geo3d_plot,
wdw_size = (1024, 768)
)
[23]:
display_image(f"{filename_view}.gif")
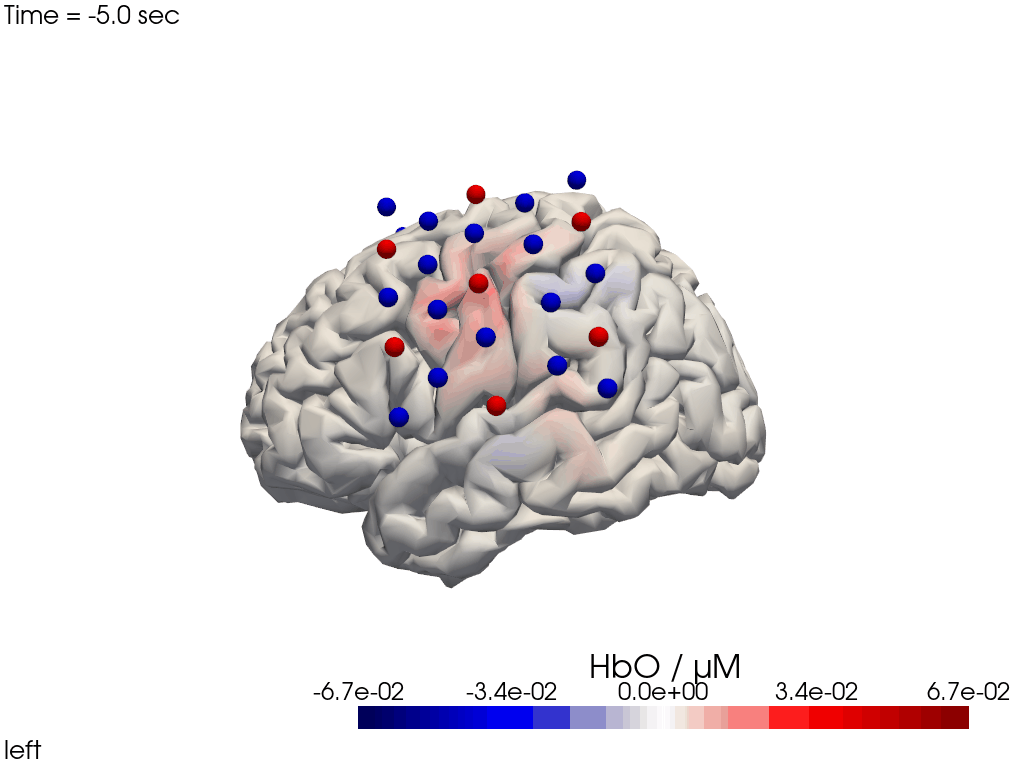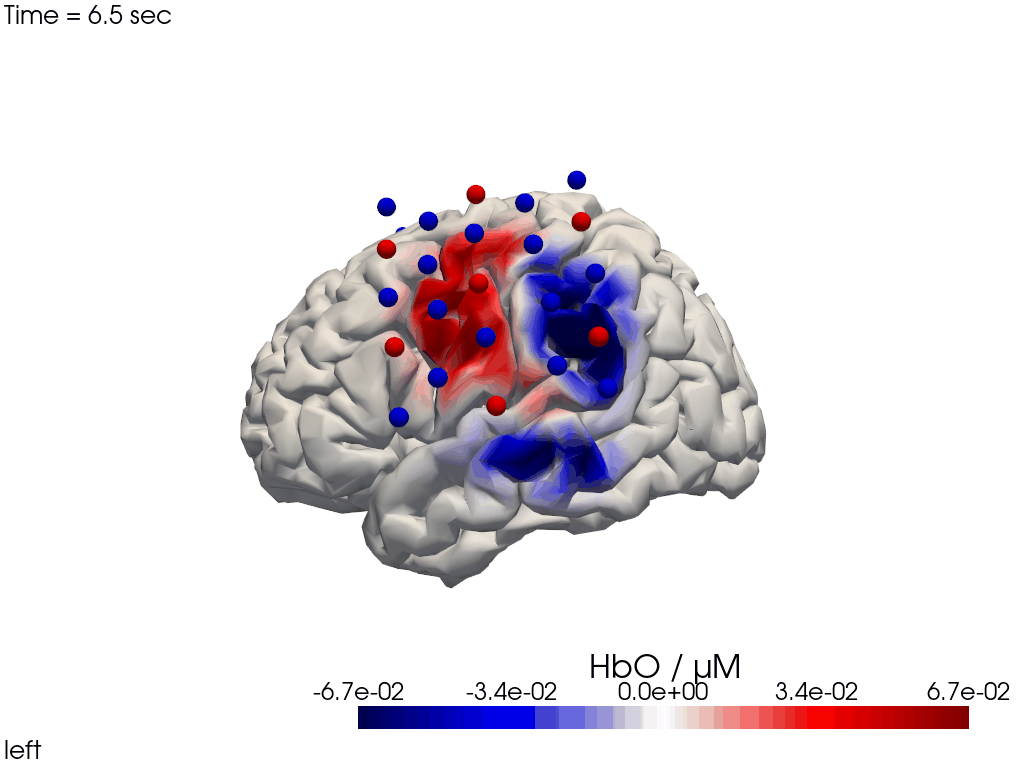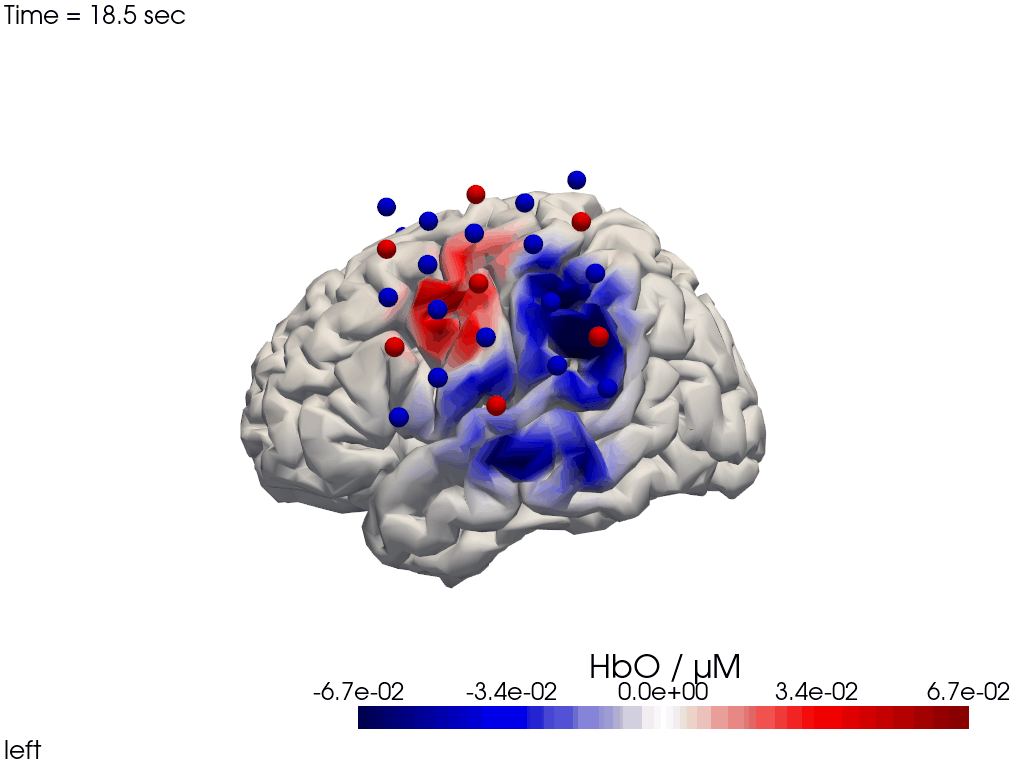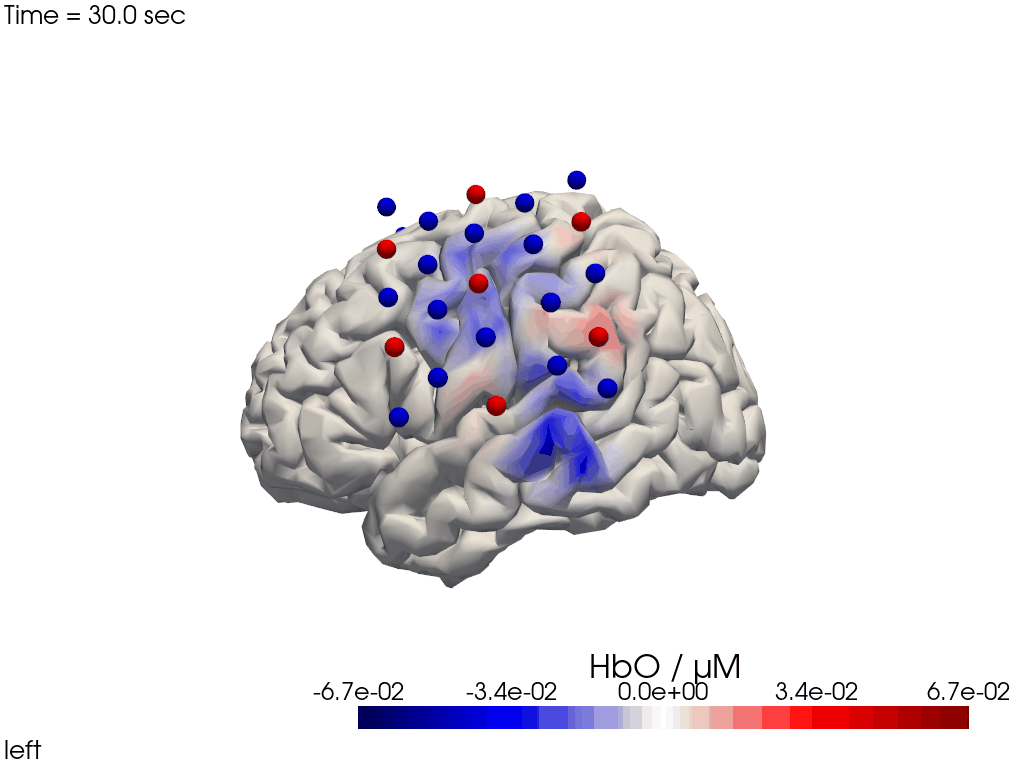
Alternatively, we can just select a single time point and plot activity as a still image at that time. Note the different file suffix (.png).
[24]:
# selects the nearest time sample at t=4s in X_ts
# note: sel does not accept quantified units
X_ts_plot = X_ts.sel(time=4, method="nearest")
filename_view = 'image_recon_view_still'
cedalion.vis.anatomy.image_recon_view(
X_ts_plot, # time series data; can be 2D (static) or 3D (dynamic)
head_ijk,
cmap='seismic',
clim=clim,
view_type='hbo_brain',
view_position='left',
title_str='HbO / µM',
filename=filename_view,
SAVE=True,
time_range=(-5,30,0.5)*units.s,
fps=6,
geo3d_plot = geo3d_plot,
wdw_size = (1024, 768)
)
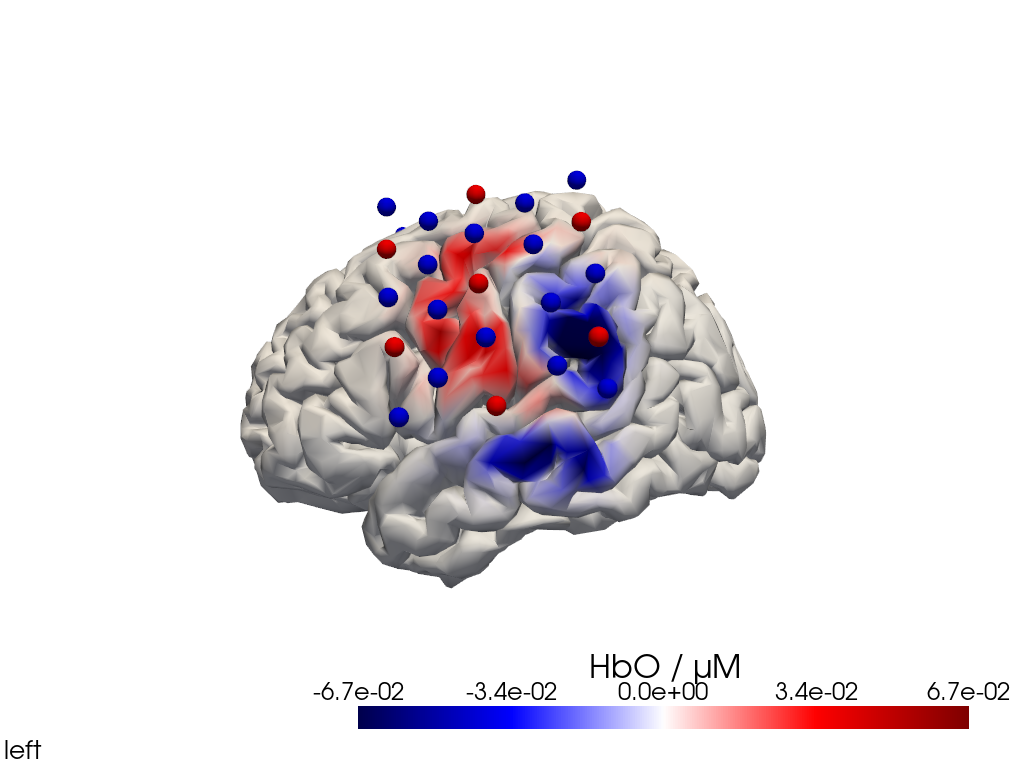
[25]:
display_image(f"{filename_view}.png")
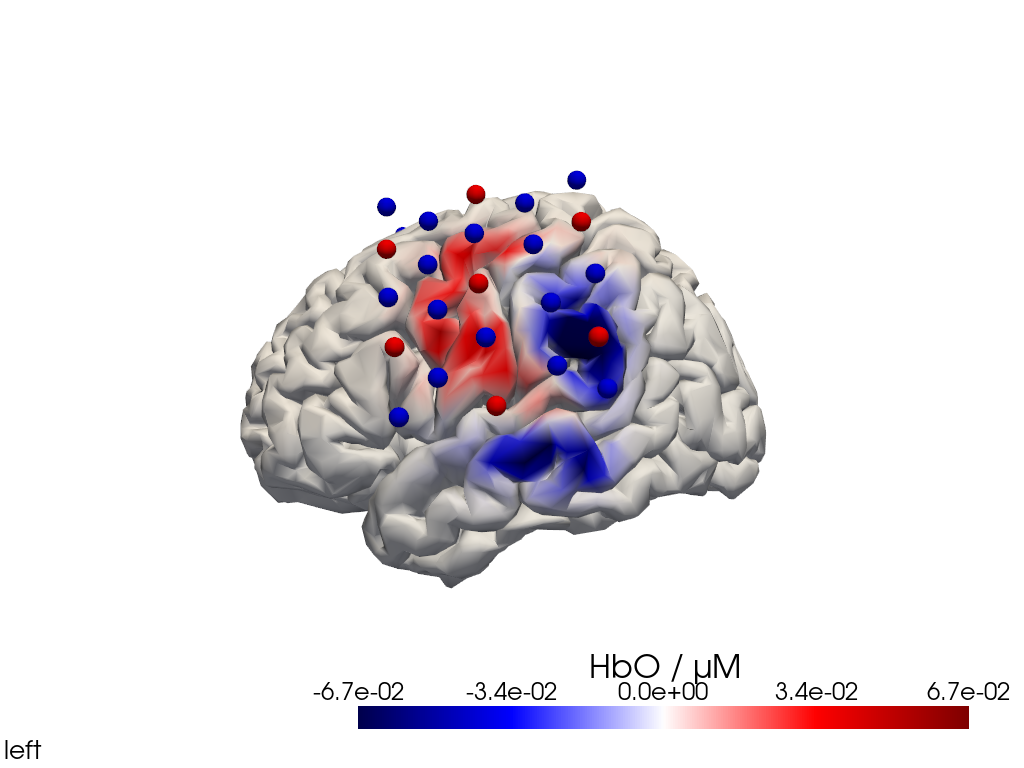
Multi-View Animations of Activitations on the Brain
The function cedalion.vis.anatomy.image_recon_multi_view shows the activity on the brain from all angles as still image or animated across time:
[26]:
filename_multiview = 'image_recon_multiview'
# prepare data
X_ts = img.sel(trial_type="FTapping/Right").rename({"reltime": "time"})
X_ts = X_ts.transpose("vertex", "chromo", "time")
scl = np.percentile(np.abs(X_ts.sel(chromo='HbO')).pint.dequantify(), 99)
clim = (-scl,scl)
cedalion.vis.anatomy.image_recon_multi_view(
X_ts, # time series data; can be 2D (static) or 3D (dynamic)
head_ijk,
cmap='seismic',
clim=clim,
view_type='hbo_brain',
title_str='HbO / µM',
filename=filename_multiview,
SAVE=True,
time_range=(-5,30,0.5)*units.s,
fps=6,
geo3d_plot = None, # geo3d_plot
wdw_size = (1024, 768)
)
[27]:
display_image(f"{filename_multiview}.gif")
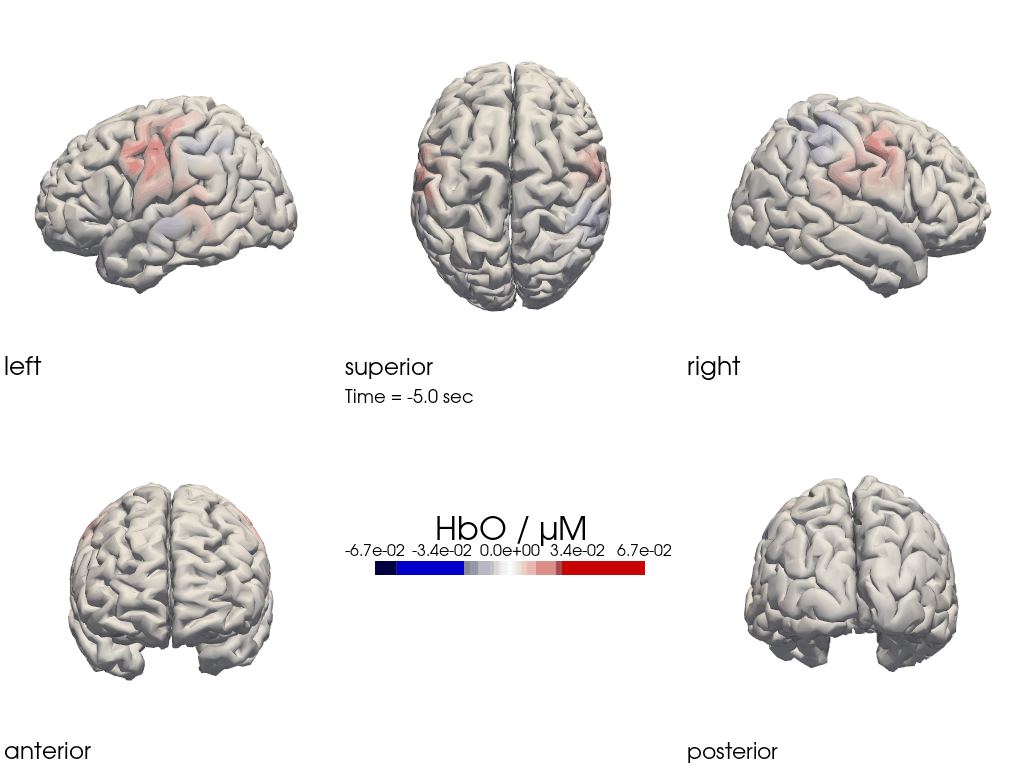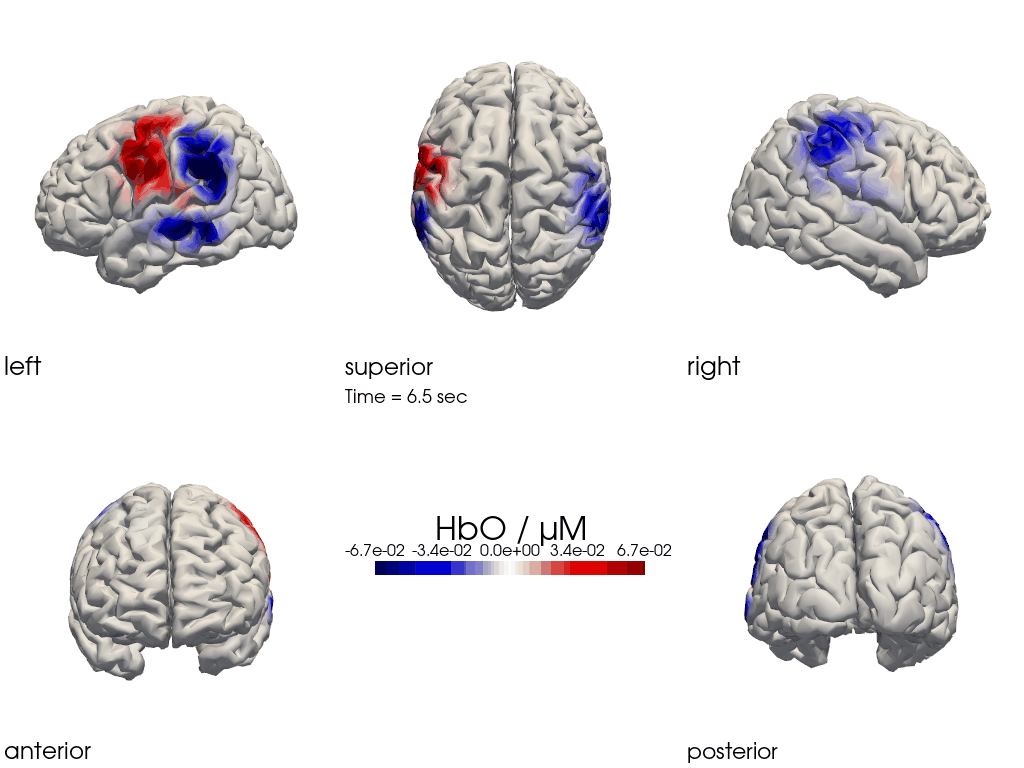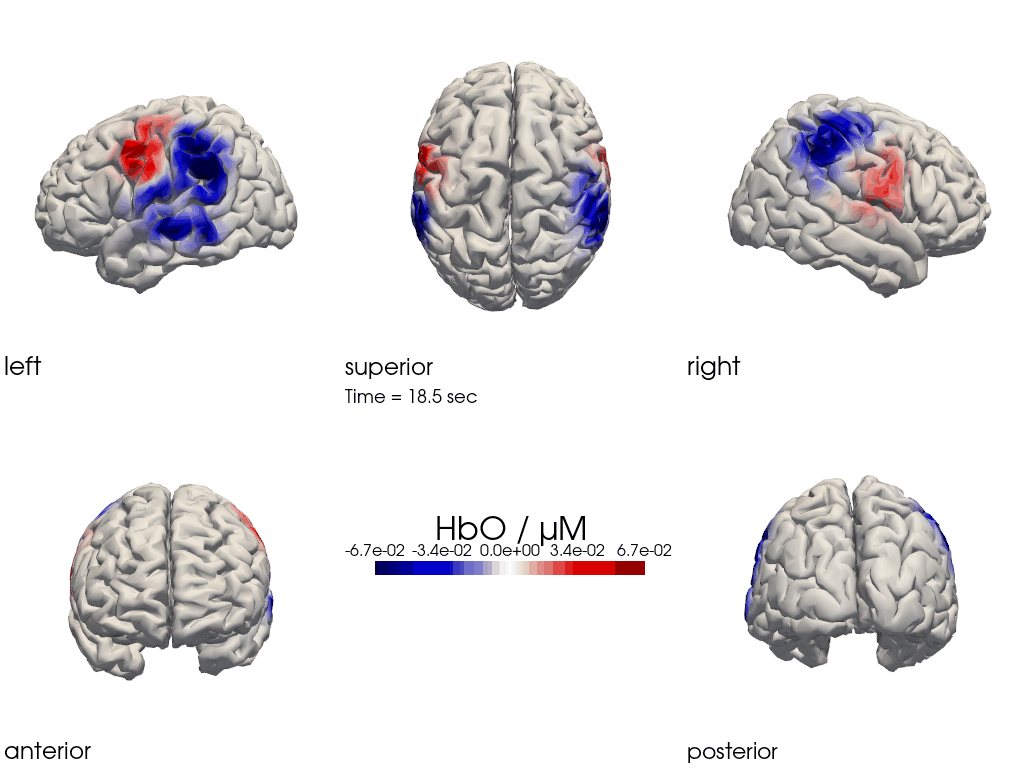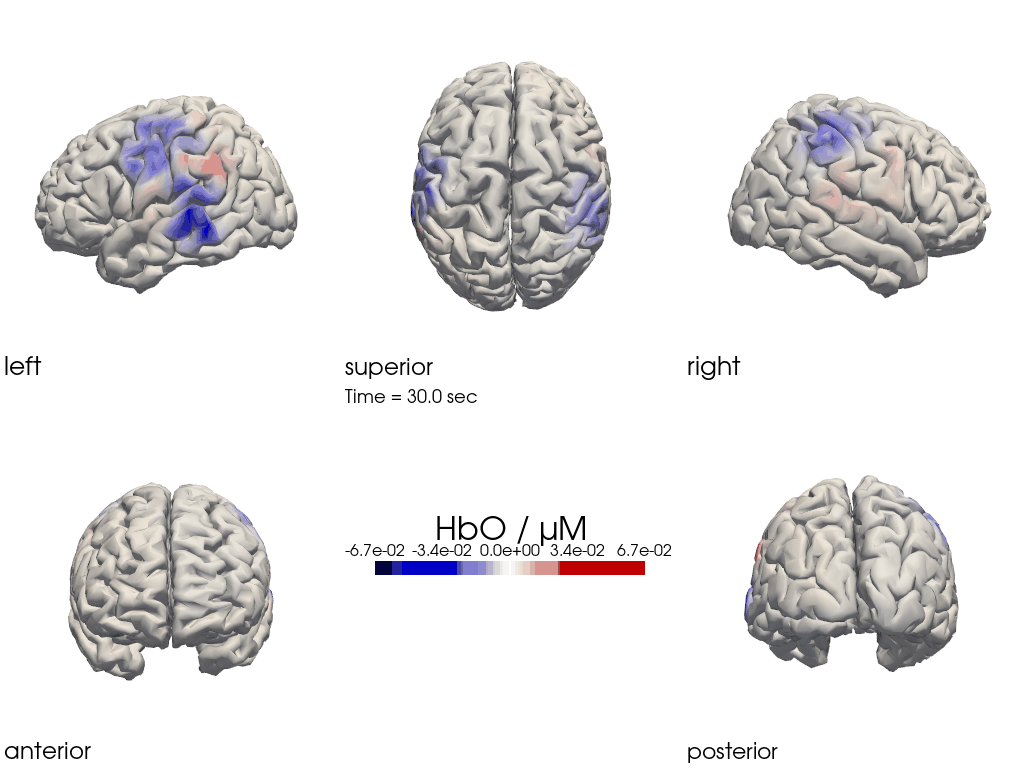
Multi-View Animations of Activitations on the Scalp
This gives us activity on the scalp after recon from all angles as still image or across time
[28]:
filename_multiview_scalp = 'image_recon_multiview_scalp'
# prepare data
X_ts = img.sel(trial_type="FTapping/Right").rename({"reltime": "time"})
X_ts = X_ts.transpose("vertex", "chromo", "time")
scl = np.percentile(np.abs(X_ts.sel(chromo='HbO')).pint.dequantify(), 99)
clim = (-scl,scl)
cedalion.vis.anatomy.image_recon_multi_view(
X_ts, # time series data; can be 2D (static) or 3D (dynamic)
head_ijk,
cmap='seismic',
clim=clim,
view_type='hbo_scalp',
title_str='HbO / uM',
filename=filename_multiview_scalp,
SAVE=True,
time_range=(-5,30,0.5)*units.s,
fps=6,
geo3d_plot = geo3d_plot,
wdw_size = (1024, 768)
)
[29]:
display_image(f"{filename_multiview_scalp}.gif")
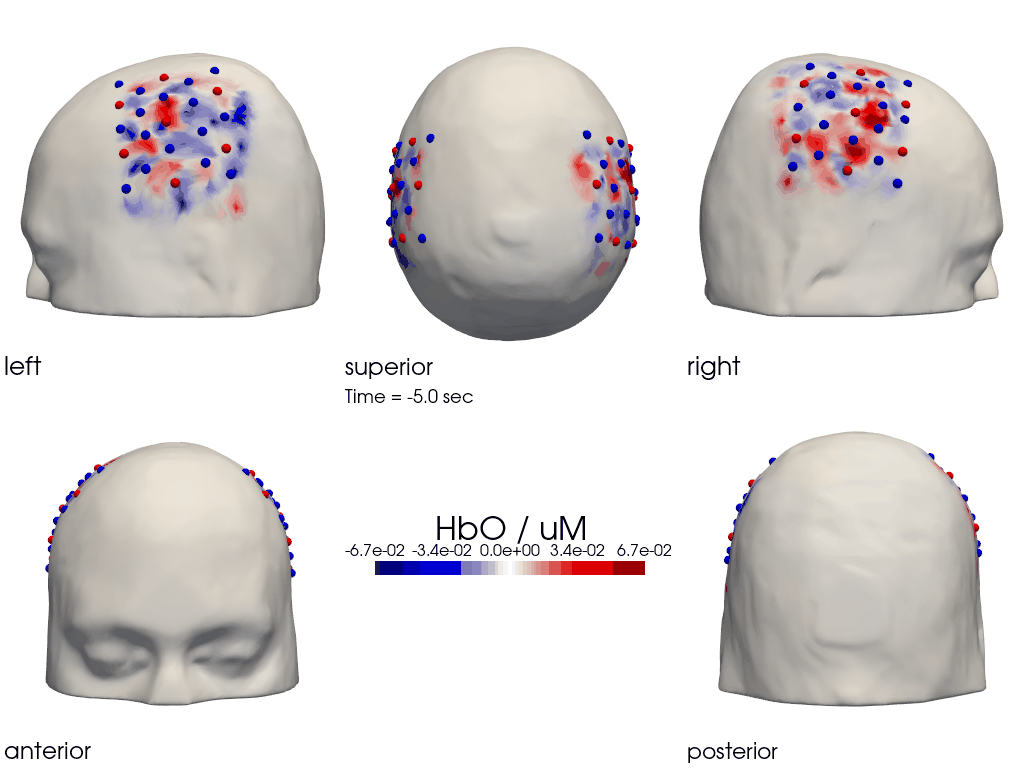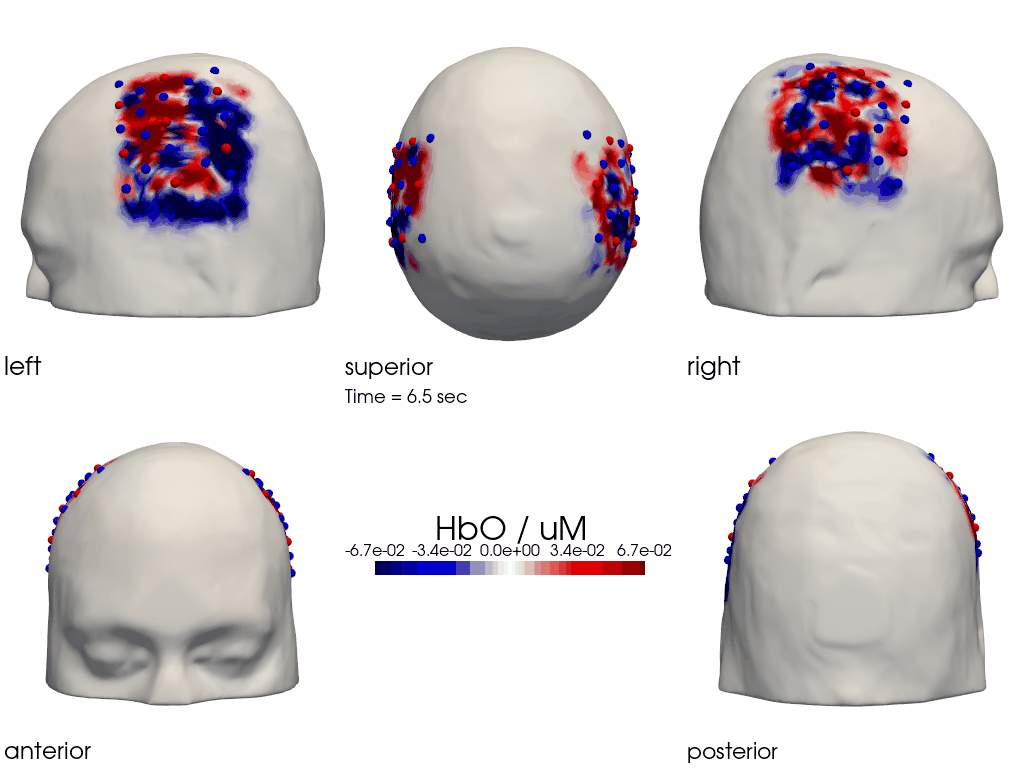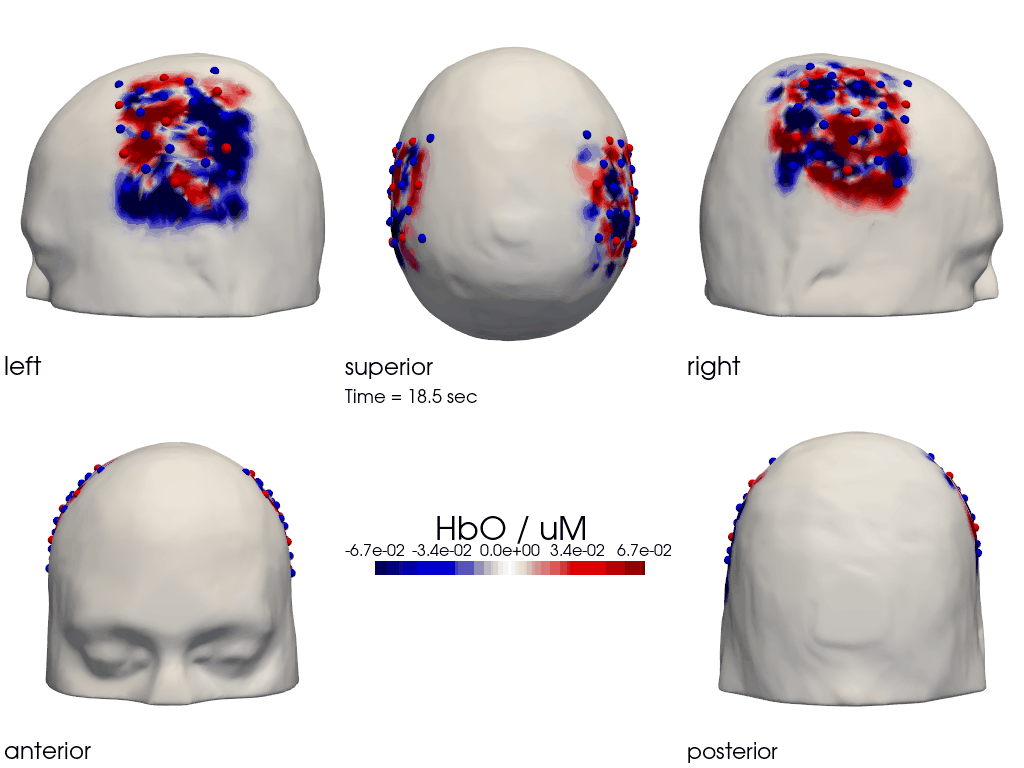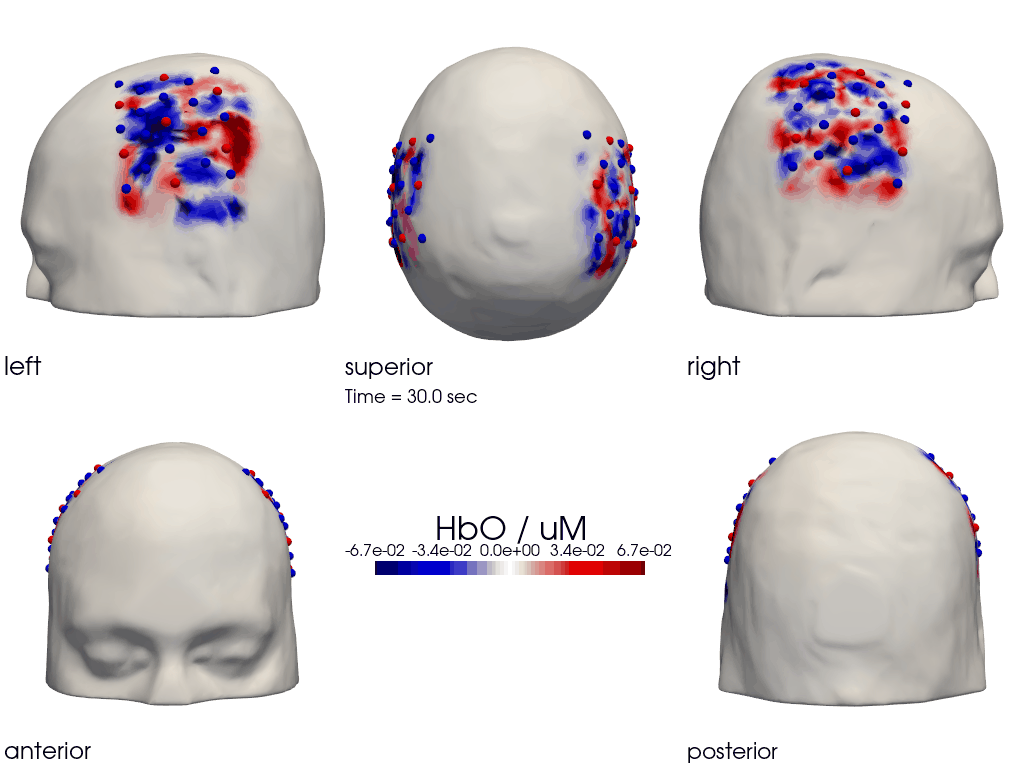
Parcel Space
The Schaefer Atlas [SKG+18] as implemented in Cedalion provides nearly 600 labels for different regions of the brain. Each vertex of the brain surface has its correspondng parcel label assigned as a coordinate.
[30]:
head_ijk.brain.vertices
[30]:
<xarray.DataArray (label: 15002, ijk: 3)> Size: 360kB [] 77.71 20.23 74.63 83.52 19.84 69.31 ... 128.0 152.7 100.5 97.28 105.3 87.73 Coordinates: * label (label) int64 120kB 0 1 2 3 4 5 ... 14997 14998 14999 15000 15001 * parcel (label) <U44 3MB 'VisCent_ExStr_8_LH' ... 'Background+FreeSurfer... Dimensions without coordinates: ijk
To obtain the average hemodynamic response in a parcel, the baseline-subtraced concentration changes of all vertices in a parcel are averaged. As baseline the first sample is used.
[31]:
# only consider brain vertices in the following.
img_brain = img.sel(vertex=img.is_brain)
# use the first sample along the reltime dimension as baseline
baseline = img_brain.isel(reltime=0)
img_brain_blsubtracted = img_brain - baseline
display(img_brain_blsubtracted.rename("img_brain_blsubtracted"))
<xarray.DataArray 'img_brain_blsubtracted' (chromo: 2, vertex: 15002,
trial_type: 2, reltime: 154)> Size: 74MB
[µM] 0.0 2.36e-12 4.763e-12 7.323e-12 ... 1.409e-11 1.376e-11 1.334e-11
Coordinates:
* chromo (chromo) <U3 24B 'HbO' 'HbR'
parcel (vertex) object 120kB 'VisCent_ExStr_8_LH' ... 'Background+Fr...
is_brain (vertex) bool 15kB True True True True ... True True True True
* reltime (reltime) float64 1kB -5.038 -4.809 -4.58 ... 29.54 29.77 30.0
* trial_type (trial_type) object 16B 'FTapping/Left' 'FTapping/Right'
Dimensions without coordinates: vertexUsing parcel labels, vertices belonging to the same brain region can be easily grouped together with the DataArray.groupby function. On these vertex groups many reduction methods can be executed. He we use the function mean() to average over vertices.
Note how the time series dimension changed from vertex to parcel:
[32]:
# average over parcels
avg_HbO = img_brain_blsubtracted.sel(chromo="HbO").groupby('parcel').mean()
avg_HbR = img_brain_blsubtracted.sel(chromo="HbR").groupby('parcel').mean()
display(avg_HbO.rename("avg_HbO"))
<xarray.DataArray 'avg_HbO' (parcel: 602, trial_type: 2, reltime: 154)> Size: 1MB
[µM] 0.0 1.002e-09 2.205e-09 3.714e-09 ... -8.357e-12 -8.706e-12 -8.967e-12
Coordinates:
chromo <U3 12B 'HbO'
* reltime (reltime) float64 1kB -5.038 -4.809 -4.58 ... 29.54 29.77 30.0
* trial_type (trial_type) object 16B 'FTapping/Left' 'FTapping/Right'
* parcel (parcel) object 5kB 'Background+FreeSurfer_Defined_Medial_Wal...The montage in this dataset covers only parts of the head. Consequently, many brain regions lack significant signal coverage due to the absence of optodes.
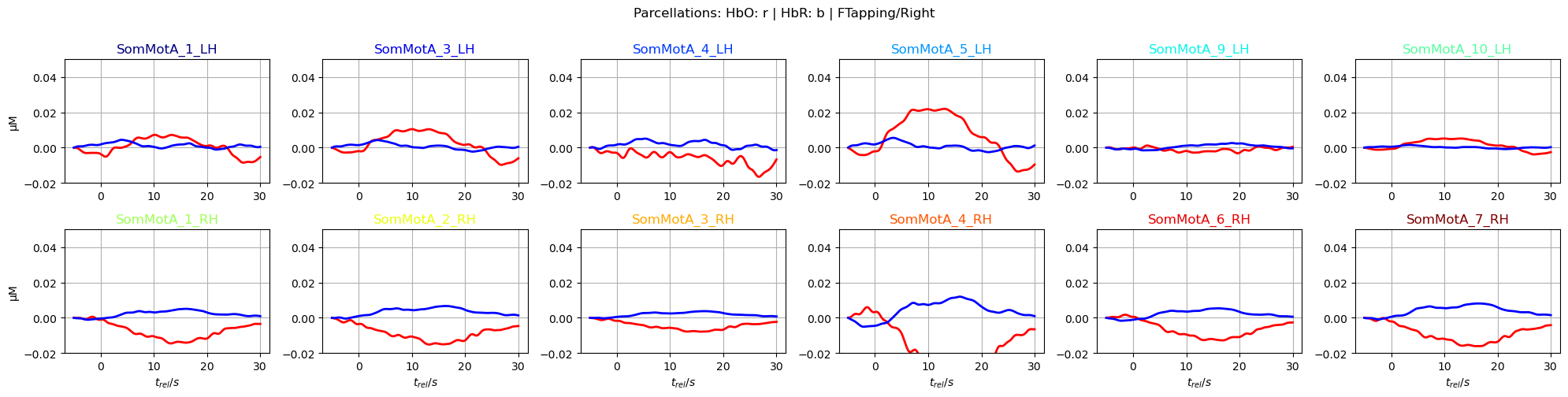
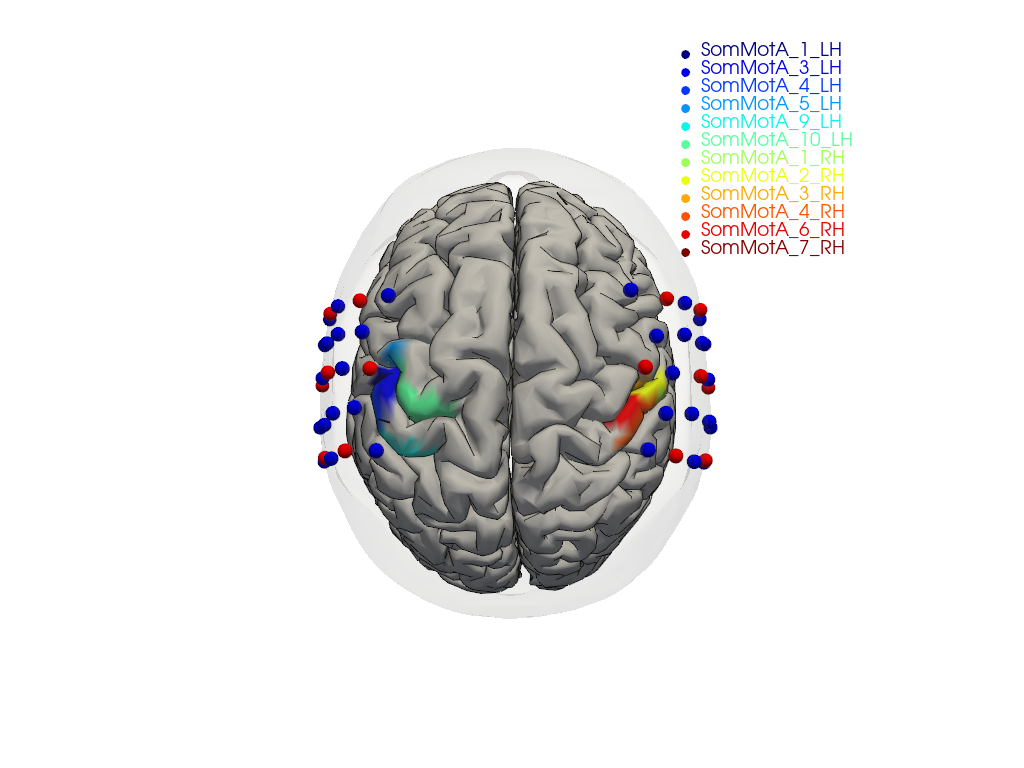
To focus on relevant regions, a subset of parcels from the somatosensory and motor regions in both hemispheres is selected.
[33]:
selected_parcels = [
"SomMotA_1_LH", "SomMotA_3_LH", "SomMotA_4_LH",
"SomMotA_5_LH", "SomMotA_9_LH", "SomMotA_10_LH",
"SomMotA_1_RH", "SomMotA_2_RH", "SomMotA_3_RH",
"SomMotA_4_RH", "SomMotA_6_RH", "SomMotA_7_RH"
]
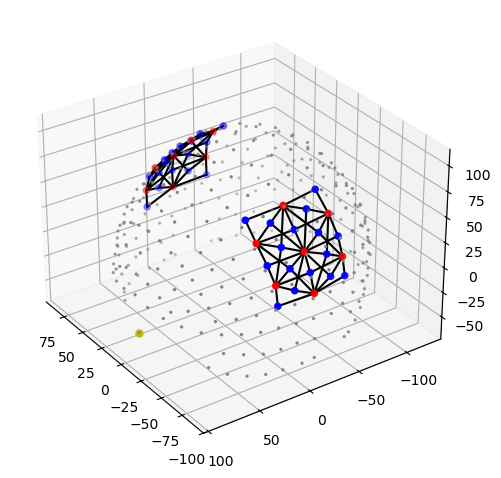
The following plot visualizes the montage and the selected parcels:
[34]:
# map parcel labels to colors
parcel_colors = {
parcel: p.cm.jet(i / (len(selected_parcels) - 1))
for i, parcel in enumerate(selected_parcels)
}
plt = pv.Plotter()
vbx.plot_surface(
plt,
head_ijk.scalp,
color="w",
opacity=.1
)
vbx.plot_surface(
plt,
head_ijk.brain,
color=np.asarray(
[
parcel_colors.get(parcel, (0.8, 0.8, 0.8, 1.))
for parcel in head_ijk.brain.vertices.parcel.values
]
),
silhouette=True,
)
vbx.plot_labeled_points(plt, geo3d_plot)
plt.add_legend(labels=parcel_colors.items(), face="o", size=(0.3, 0.3))
vbx.camera_at_cog(plt, head_ijk.brain, rpos=(0, 0, 1), up=(0, 1, 0), fit_scene=True)
plt.show()
Plot averaged time traces in each parcel for the ‘FTapping/Right’ and ‘FTapping/Left’ conditions:
[35]:
f,ax = p.subplots(2,6, figsize=(20,5))
ylims = -0.02, 0.05
ax = ax.flatten()
for i_par, par in enumerate(selected_parcels):
ax[i_par].plot(avg_HbO.sel(parcel = par, trial_type = "FTapping/Right").reltime, avg_HbO.sel(parcel = par, trial_type = "FTapping/Right"), "r", lw=2, ls='-')
ax[i_par].plot(avg_HbR.sel(parcel = par, trial_type = "FTapping/Right").reltime, avg_HbR.sel(parcel = par, trial_type = "FTapping/Right"), "b", lw=2, ls='-')
ax[i_par].grid(1)
ax[i_par].set_title(par, color=parcel_colors[par])
ax[i_par].set_ylim(*ylims)
if i_par % 6 == 0:
ax[i_par].set_ylabel("µM")
if i_par >=6:
ax[i_par].set_xlabel("$t_{rel} / s$")
p.suptitle("Parcellations: HbO: r | HbR: b | FTapping/Right", y=1)
p.tight_layout()
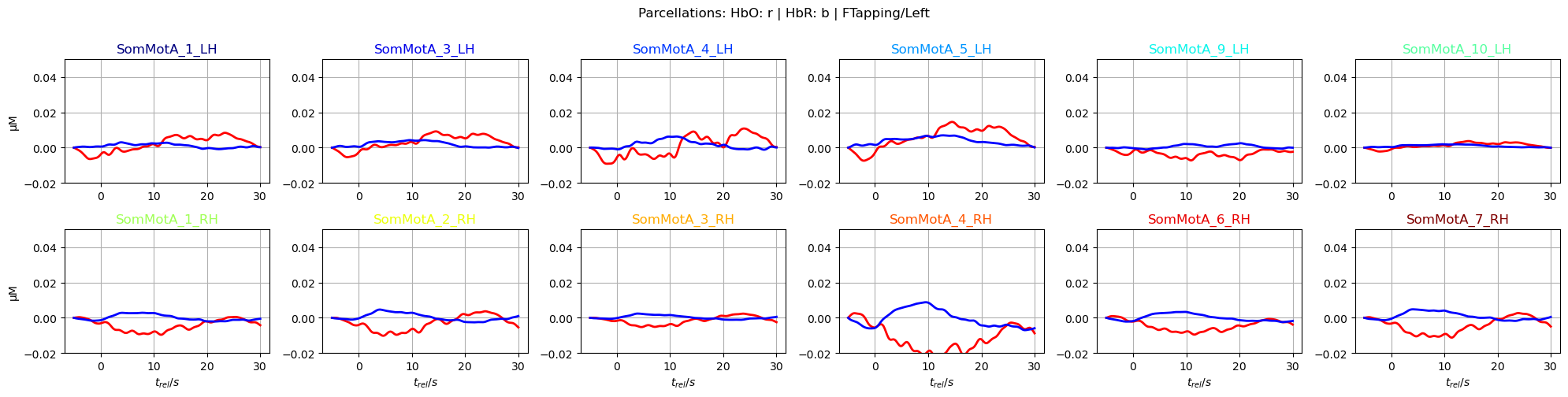
f,ax = p.subplots(2,6, figsize=(20,5))
ax = ax.flatten()
for i_par, par in enumerate(selected_parcels):
ax[i_par].plot(avg_HbO.sel(parcel = par, trial_type = "FTapping/Left").reltime, avg_HbO.sel(parcel = par, trial_type = "FTapping/Left"), "r", lw=2, ls='-')
ax[i_par].plot(avg_HbR.sel(parcel = par, trial_type = "FTapping/Left").reltime, avg_HbR.sel(parcel = par, trial_type = "FTapping/Left"), "b", lw=2, ls='-')
ax[i_par].grid(1)
ax[i_par].set_title(par, color=parcel_colors[par])
ax[i_par].set_ylim(*ylims)
if i_par % 6 == 0:
ax[i_par].set_ylabel("µM")
if i_par >=6:
ax[i_par].set_xlabel("$t_{rel} / s$")
p.suptitle("Parcellations: HbO: r | HbR: b | FTapping/Left", y=1)
p.tight_layout()