Head model fiducials and landmarks
Cedalion ships with segmentations as well as brain and scalp surfaces for the Colin27 and ICBM-152 heads.
This notebook documents the source of the fiducial landmarks and compares the outputs of the landmark builder which we distribute together with the head models.
[1]:
# This cells setups the environment when executed in Google Colab.
try:
import google.colab
!curl -s https://raw.githubusercontent.com/ibs-lab/cedalion/dev/scripts/colab_setup.py -o colab_setup.py
# Select branch with --branch "branch name" (default is "dev")
%run colab_setup.py
except ImportError:
pass
[2]:
import cedalion
import cedalion.dataclasses as cdc
import cedalion.data
import cedalion.geometry.landmarks
import cedalion.geometry.segmentation
import cedalion.io
import cedalion.vis.blocks as vbx
import cedalion.dot
import cedalion.xrutils as xrutils
import matplotlib.pyplot as p
import numpy as np
import pyvista
import xarray as xr
pyvista.set_jupyter_backend("static")
#pyvista.set_jupyter_backend("server")
Utility function
[3]:
def compare_landmarks(scalp, landmarks, reference):
fiducial_labels = ["Nz", "Iz", "LPA", "RPA", "Cz"]
plt = pyvista.Plotter()
vbx.plot_surface(plt, scalp, opacity=0.7)
vbx.plot_labeled_points(plt, reference, color="y")
vbx.plot_labeled_points(plt, landmarks, color="g")
vbx.plot_labeled_points(plt, landmarks.loc[fiducial_labels], color="r")
plt.show()
common_labels = list(set(reference.label.values) & set(landmarks.label.values))
diffs = xrutils.norm(
landmarks.sel(label=landmarks.label.isin(common_labels))
- reference.sel(label=reference.label.isin(common_labels)),
dim="mni",
)
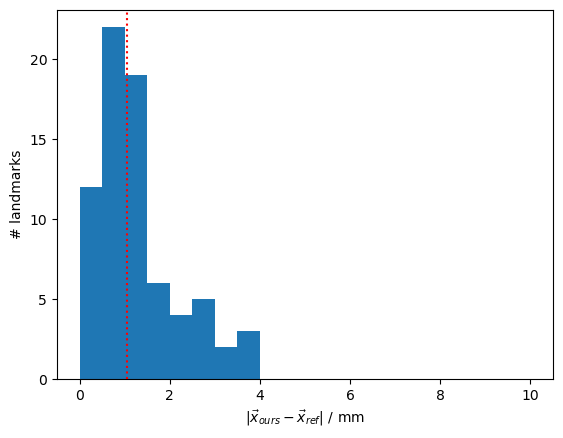
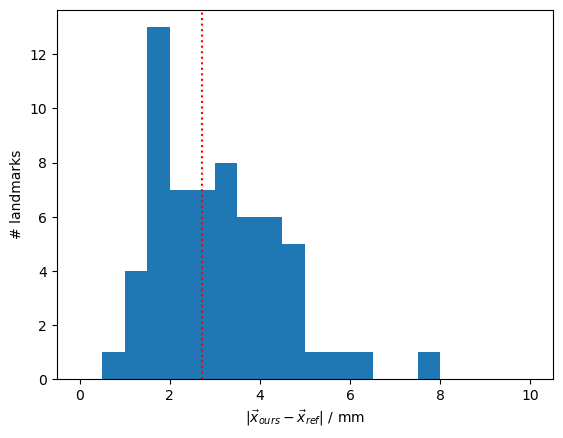
diffs = diffs.pint.to("mm").pint.dequantify()
median_diff = np.median(diffs).item()
p.figure()
p.hist(diffs, np.arange(0, 10.5, 0.5))
p.axvline(median_diff, c="r", ls=":")
p.xlabel(r"$|\vec{x}_{ours} - \vec{x}_{ref}|$ / mm")
p.ylabel(r"# landmarks")
print(f"The median distance between landmarks is {median_diff:.1f} mm.")
Colin 27
Load Colin27 head model
[4]:
colin_ijk = cedalion.dot.get_standard_headmodel("colin27")
colin_ras = colin_ijk.apply_transform(colin_ijk.t_ijk2ras)
Load fieldtrip 10-5 coordinates:
[5]:
colin_fieldtrip = cedalion.data.get_fieldtrip_colin27_landmarks()
colin_fieldtrip = colin_fieldtrip.points.set_crs("mni")
colin_fieldtrip
[5]:
<xarray.DataArray (label: 346, mni: 3)> Size: 8kB
<Quantity([[-8.60761e+01 -1.99897e+01 -4.79860e+01]
[ 8.57939e+01 -2.00093e+01 -4.80310e+01]
[ 8.30000e-03 8.68110e+01 -3.99830e+01]
...
[ 8.57939e+01 -4.50093e+01 -6.80310e+01]
[-8.60761e+01 -2.49897e+01 -6.79860e+01]
[ 8.57939e+01 -2.50093e+01 -6.80310e+01]], 'millimeter')>
Coordinates:
* label (label) <U6 8kB 'LPA' 'RPA' 'Nz' 'Fp1' ... 'M1' 'M2' 'A1' 'A2'
type (label) object 3kB PointType.LANDMARK ... PointType.LANDMARK
Dimensions without coordinates: mniVisualize fieldtrip coordinates and the Colin27 scalp surface that ships with Cedalion. Note tha Nz floats above the surface.
[6]:
plt = pyvista.Plotter()
vbx.plot_surface(plt, colin_ras.scalp, opacity=.7)
vbx.plot_labeled_points(plt, colin_fieldtrip, color="y")
plt.show()
Select the five fiducicals, with Nz snapped to the scalp.
[7]:
colin_fiducials = xr.concat(
(
colin_fieldtrip.loc[["Iz", "LPA", "RPA", "Cz"]],
colin_ras.scalp.snap(colin_fieldtrip.loc[["Nz"]]),
),
dim="label",
)
colin_fiducials
[7]:
<xarray.DataArray (label: 5, mni: 3)> Size: 120B
<Quantity([[ 4.5000000e-03 -1.1856500e+02 -2.3078000e+01]
[-8.6076100e+01 -1.9989700e+01 -4.7986000e+01]
[ 8.5793900e+01 -2.0009300e+01 -4.8031000e+01]
[ 4.0090000e-01 -9.1670000e+00 1.0024400e+02]
[-6.7919500e-01 8.3259347e+01 -3.9408527e+01]], 'millimeter')>
Coordinates:
* label (label) <U6 120B 'Iz' 'LPA' 'RPA' 'Cz' 'Nz'
type (label) object 40B PointType.LANDMARK ... PointType.LANDMARK
Dimensions without coordinates: mniConstruct remaining landmarks:
[8]:
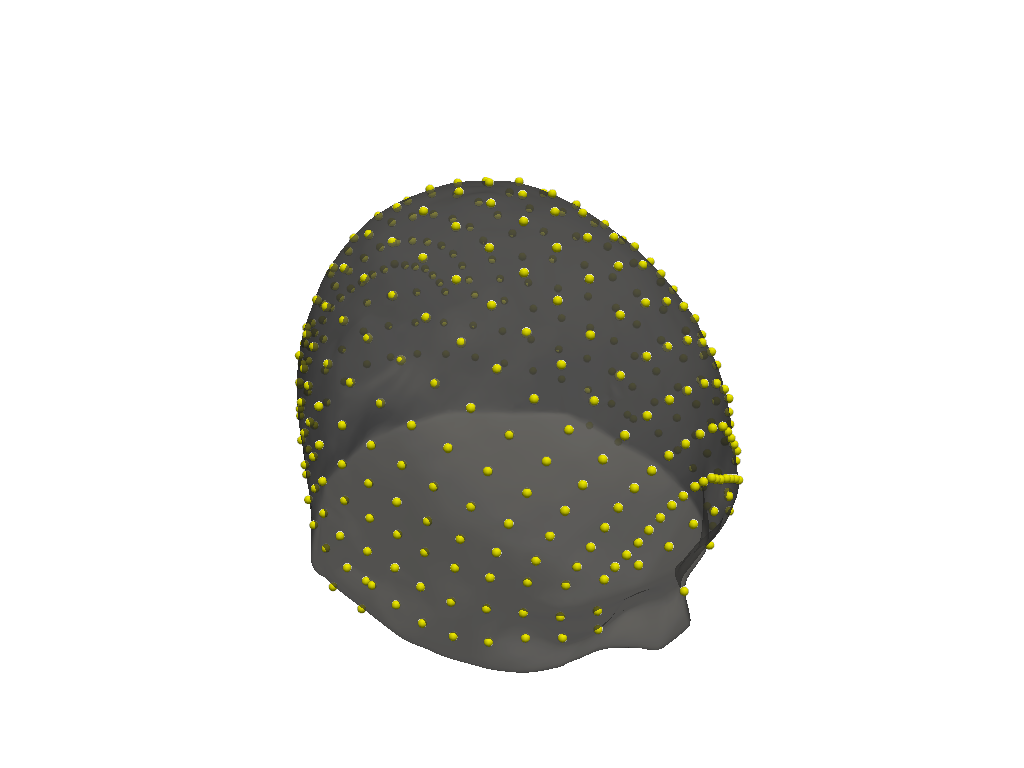
lmbuilder = cedalion.geometry.landmarks.LandmarksBuilder1010(colin_ras.scalp, colin_fiducials)
colin_all_landmarks = lmbuilder.build()
lmbuilder.plot()
/home/eike/Projekte/ibslab/30_dev/cedalion/src/cedalion/geometry/landmarks.py:263: UserWarning: WIP: distance calculation around ears
warnings.warn("WIP: distance calculation around ears")

Compare constructed landmarks to fieldtrip reference
[9]:
compare_landmarks(colin_ras.scalp, colin_all_landmarks, colin_fieldtrip)
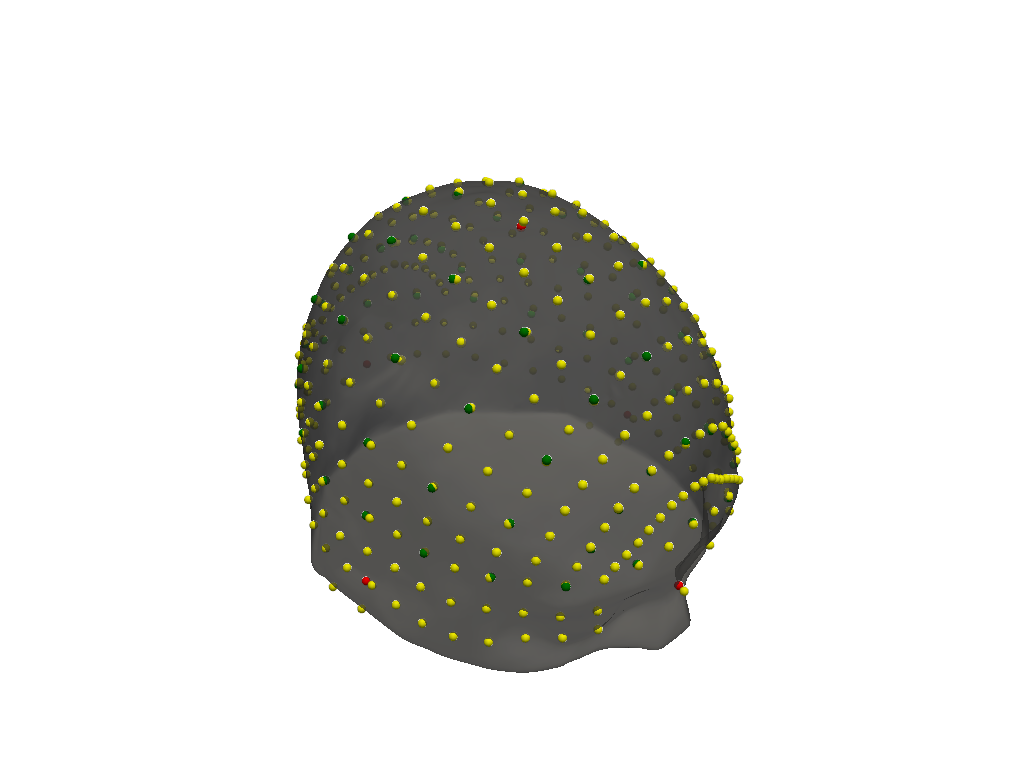
The median distance between landmarks is 1.0 mm.
ICBM-152
The fiducials and reference landmarks for the ICBM-152 head were adopted from Cutini et al.
Translate data from the paper to labeled points:
[10]:
def get_cutini_icbm_landmarks():
# from the supplementary table of Cutini et al., 2010
# doi: 10.1016/j.neuroimage.2010.09.030
cutini_icbm_coords = """
Fp1 -28 83 -5
Fp2 31 84 -5
Fz 0 58 66
F3 -50 55 39
F4 50 55 41
F7 -68 41 -12
F8 69 41 -12
Cz 0 -9 100
C3 -66 -11 63
C4 69 -11 63
T7 -80 -16 -10
T8 81 -17 -10
Pz 0 -82 78
P3 -52 -80 54
P4 53 -80 54
P7 -71 -72 -5
P8 70 -72 -5
O1 -26 -111 3
O2 27 -109 3
Fpz 0 88 -1
AFz 0 79 37
AF3 -40 72 24
AF4 40 73 24
AF7 -53 66 -9
AF8 54 65 -9
F1 -30 60 56
F2 29 59 59
F5 -63 47 13
F6 64 47 15
FCz 0 26 87
FC1 -36 28 76
FC2 35 27 77
FC3 -60 25 52
FC4 62 24 54
FC5 -74 19 18
FC6 75 20 21
FT7 -75 13 -14
FT8 78 13 -15
C1 -38 -8 89
C2 38 -10 89
C5 -80 -15 26
C6 81 -14 26
CPz 0 -49 99
CP1 -37 -48 91
CP2 37 -51 90
CP3 -65 -50 66
CP4 65 -51 64
CP5 -79 -47 30
CP6 78 -48 29
TP7 -80 -45 -8
TP8 80 -45 -9
P1 -28 -83 74
P2 30 -83 73
P5 -66 -79 26
P6 65 -77 26
POz 0 -103 46
PO3 -37 -101 37
PO4 39 -100 37
PO7 -54 -97 0
PO8 52 -97 0
Oz 0 -115 10
"""
labels = []
coords = []
for line in cutini_icbm_coords.splitlines():
line = line.strip()
if not line:
continue
fields = line.split()
labels.append(fields[0])
coords.append(np.asarray([float(i) for i in fields[1:]]))
return cdc.build_labeled_points(
np.asarray(coords), crs="mni", units="mm", labels=labels
)
def get_cutini_fiducials():
# from Cutini et al., 2010, Table 1
# doi: 10.1016/j.neuroimage.2010.09.030
return cdc.build_labeled_points(
np.asarray(
[
[0.0, 84.0, -43.0],
[0.0, -114.0, -30.0],
[-75.09, -19.49, -47.98],
[76, -19.45, -47.7],
[0.0, -9.0, 100.0],
]
),
crs="mni",
units="mm",
labels=["Nz", "Iz", "LPA", "RPA", "Cz"],
)
[11]:
icbm_ijk = cedalion.dot.get_standard_headmodel("icbm152")
icbm_ras = icbm_ijk.apply_transform(icbm_ijk.t_ijk2ras)
[12]:
plt = pyvista.Plotter()
vbx.plot_surface(plt, icbm_ras.scalp, opacity=.7)
vbx.plot_labeled_points(plt, get_cutini_icbm_landmarks(), color="y")
vbx.plot_labeled_points(plt, get_cutini_fiducials(), color="r")
plt.show()

[13]:
icbm_fiducials = icbm_ras.scalp.snap(get_cutini_fiducials())
icbm_fiducials = get_cutini_fiducials()
[14]:
lmbuilder = cedalion.geometry.landmarks.LandmarksBuilder1010(icbm_ras.scalp, icbm_fiducials)
icbm_all_landmarks = lmbuilder.build()
lmbuilder.plot()
/home/eike/Projekte/ibslab/30_dev/cedalion/src/cedalion/geometry/landmarks.py:263: UserWarning: WIP: distance calculation around ears
warnings.warn("WIP: distance calculation around ears")

[15]:
compare_landmarks(icbm_ras.scalp, icbm_all_landmarks, get_cutini_icbm_landmarks())

The median distance between landmarks is 2.7 mm.
Export landmarks
[16]:
#cedalion.io.probe_geometry.save_mrk_json("colin.mrk.json", icbm_all_landmarks, "mni")
#cedalion.io.probe_geometry.save_mrk_json("icbm.mrk.json", icbm_all_landmarks, "mni")