Storing estimated HRFs in snirf files
This notebook estimates the HRF in a finger-tapping experiment by blockaveraging and then stores the result in a snirf file.
[1]:
# This cells setups the environment when executed in Google Colab.
try:
import google.colab
!curl -s https://raw.githubusercontent.com/ibs-lab/cedalion/dev/scripts/colab_setup.py -o colab_setup.py
# Select branch with --branch "branch name" (default is "dev")
%run colab_setup.py
except ImportError:
pass
[2]:
from pathlib import Path
import tempfile
import matplotlib.pyplot as p
import numpy as np
import xarray as xr
from snirf import Snirf
import cedalion
import cedalion.data
import cedalion.io
import cedalion.nirs
from cedalion import units
xr.set_options(display_max_rows=3, display_values_threshold=50, display_expand_data=False)
np.set_printoptions(precision=4)
Load a finger-tapping dataset
For this demo we load an example finger-tapping recording through cedalion.data.get_fingertapping. The file contains a single NIRS element with one block of raw amplitude data.
[3]:
rec = cedalion.data.get_fingertapping()
[4]:
# Rename events
rec.stim.cd.rename_events( {
"1.0" : "control",
"2.0" : "Tapping/Left",
"3.0" : "Tapping/Right"
})
Calculate concentrations
[5]:
dpf = xr.DataArray([6, 6], dims="wavelength", coords={"wavelength" : rec["amp"].wavelength})
rec["od"] = - np.log( rec["amp"] / rec["amp"].mean("time") )
rec["conc"] = cedalion.nirs.cw.beer_lambert(rec["amp"], rec.geo3d, dpf)
Frequency filtering and splitting into epochs
[6]:
rec["conc_freqfilt"] = rec["conc"].cd.freq_filter(fmin=0.02, fmax=0.5, butter_order=4)
cf_epochs = rec["conc_freqfilt"].cd.to_epochs(
rec.stim, # stimulus dataframe
["Tapping/Left", "Tapping/Right"], # select events
before=5 * units.s, # seconds before stimulus
after=20 * units.s, # seconds after stimulus
)
Blockaveraging to estimate the HRFs
[7]:
# calculate baseline
baseline = cf_epochs.sel(reltime=(cf_epochs.reltime < 0)).mean("reltime")
# subtract baseline
epochs_blcorrected = cf_epochs - baseline
# group trials by trial_type. For each group individually average the epoch dimension
rec["hrf_blockaverage"] = epochs_blcorrected.groupby("trial_type").mean("epoch")
display(rec["hrf_blockaverage"])
<xarray.DataArray (trial_type: 2, chromo: 2, channel: 28, reltime: 198)> Size: 177kB
[µM] -0.05277 -0.05263 -0.05258 -0.05263 ... -0.005955 -0.007018 -0.00833
Coordinates: (3/6)
* reltime (reltime) float64 2kB -5.12 -4.992 -4.864 ... 19.84 19.97 20.1
* chromo (chromo) <U3 24B 'HbO' 'HbR'
... ...
* trial_type (trial_type) object 16B 'Tapping/Left' 'Tapping/Right'Store HRFs
[8]:
tmpdir = tempfile.TemporaryDirectory()
snirf_fname = str(Path(tmpdir.name) / "test.snirf")
[9]:
cedalion.io.snirf.write_snirf(snirf_fname, rec)
Inspect snirf file
[10]:
s = Snirf(snirf_fname)
[11]:
display(s.nirs)
display(s.nirs[0].data)
<iterable of 1 <class 'snirf.pysnirf2.NirsElement'>>
<iterable of 5 <class 'snirf.pysnirf2.DataElement'>>
Stim DataFrame
[12]:
display(s.nirs[0].stim)
<iterable of 4 <class 'snirf.pysnirf2.StimElement'>>
[13]:
cedalion.io.snirf.stim_to_dataframe(s.nirs[0].stim)
[13]:
| onset | duration | value | trial_type | |
|---|---|---|---|---|
| 0 | 61.824 | 5.0 | 1.0 | control |
| 1 | 87.296 | 5.0 | 1.0 | control |
| 2 | 181.504 | 5.0 | 1.0 | control |
| 3 | 275.712 | 5.0 | 1.0 | control |
| 4 | 544.640 | 5.0 | 1.0 | control |
| ... | ... | ... | ... | ... |
| 87 | 2234.112 | 5.0 | 1.0 | Tapping/Right |
| 88 | 2334.848 | 5.0 | 1.0 | Tapping/Right |
| 89 | 2362.240 | 5.0 | 1.0 | Tapping/Right |
| 90 | 2677.504 | 5.0 | 1.0 | Tapping/Right |
| 91 | 2908.032 | 5.0 | 1.0 | Tapping/Right |
92 rows × 4 columns
MeasurementList DataFrame
[14]:
for i_data in range(len(s.nirs[0].data)):
df_ml = cedalion.io.snirf.measurement_list_to_dataframe(
s.nirs[0].data[i_data].measurementList, drop_none=True
)
display(df_ml.head(3))
display(df_ml.tail(3))
| sourceIndex | detectorIndex | wavelengthIndex | dataType | dataUnit | |
|---|---|---|---|---|---|
| 0 | 1 | 1 | 1 | 1 | volt |
| 1 | 1 | 1 | 2 | 1 | volt |
| 2 | 1 | 2 | 1 | 1 | volt |
| sourceIndex | detectorIndex | wavelengthIndex | dataType | dataUnit | |
|---|---|---|---|---|---|
| 53 | 8 | 8 | 2 | 1 | volt |
| 54 | 8 | 16 | 1 | 1 | volt |
| 55 | 8 | 16 | 2 | 1 | volt |
| sourceIndex | detectorIndex | wavelengthIndex | dataType | dataUnit | dataTypeLabel | |
|---|---|---|---|---|---|---|
| 0 | 1 | 1 | 1 | 99999 | dimensionless | dOD |
| 1 | 1 | 1 | 2 | 99999 | dimensionless | dOD |
| 2 | 1 | 2 | 1 | 99999 | dimensionless | dOD |
| sourceIndex | detectorIndex | wavelengthIndex | dataType | dataUnit | dataTypeLabel | |
|---|---|---|---|---|---|---|
| 53 | 8 | 8 | 2 | 99999 | dimensionless | dOD |
| 54 | 8 | 16 | 1 | 99999 | dimensionless | dOD |
| 55 | 8 | 16 | 2 | 99999 | dimensionless | dOD |
| sourceIndex | detectorIndex | dataType | dataUnit | dataTypeLabel | |
|---|---|---|---|---|---|
| 0 | 1 | 1 | 99999 | micromolar | HbO |
| 1 | 1 | 1 | 99999 | micromolar | HbR |
| 2 | 1 | 2 | 99999 | micromolar | HbO |
| sourceIndex | detectorIndex | dataType | dataUnit | dataTypeLabel | |
|---|---|---|---|---|---|
| 53 | 8 | 8 | 99999 | micromolar | HbR |
| 54 | 8 | 16 | 99999 | micromolar | HbO |
| 55 | 8 | 16 | 99999 | micromolar | HbR |
| sourceIndex | detectorIndex | dataType | dataUnit | dataTypeLabel | |
|---|---|---|---|---|---|
| 0 | 1 | 1 | 99999 | micromolar | HbO |
| 1 | 1 | 1 | 99999 | micromolar | HbR |
| 2 | 1 | 2 | 99999 | micromolar | HbO |
| sourceIndex | detectorIndex | dataType | dataUnit | dataTypeLabel | |
|---|---|---|---|---|---|
| 53 | 8 | 8 | 99999 | micromolar | HbR |
| 54 | 8 | 16 | 99999 | micromolar | HbO |
| 55 | 8 | 16 | 99999 | micromolar | HbR |
| sourceIndex | detectorIndex | dataType | dataUnit | dataTypeLabel | dataTypeIndex | |
|---|---|---|---|---|---|---|
| 0 | 1 | 1 | 99999 | micromolar | HRF HbO | 3 |
| 1 | 1 | 1 | 99999 | micromolar | HRF HbR | 3 |
| 2 | 1 | 2 | 99999 | micromolar | HRF HbO | 3 |
| sourceIndex | detectorIndex | dataType | dataUnit | dataTypeLabel | dataTypeIndex | |
|---|---|---|---|---|---|---|
| 109 | 8 | 8 | 99999 | micromolar | HRF HbR | 4 |
| 110 | 8 | 16 | 99999 | micromolar | HRF HbO | 4 |
| 111 | 8 | 16 | 99999 | micromolar | HRF HbR | 4 |
[15]:
s.close()
Read HRFs from snirf file
Note: read_snirf names the time dimension time whereas in blockaverage it was called reltime. Need to agree on a convention.
[16]:
hrf_recs = cedalion.io.read_snirf(snirf_fname)
display(hrf_recs)
[<Recording | timeseries: ['amp', 'od', 'conc', 'conc_02', 'hrf_conc'], masks: [], stim: ['control', '15.0', 'Tapping/Left', 'Tapping/Right'], aux_ts: [], aux_obj: []>]
[17]:
# the name of the stored blockaverages derives from the datatype (HRF) and the type of data (concentration)
read_blockaverage = hrf_recs[0]["hrf_conc"]
display(read_blockaverage)
<xarray.DataArray (channel: 28, chromo: 2, trial_type: 2, time: 198)> Size: 177kB
[µM] -0.05277 -0.05263 -0.05258 -0.05263 ... -0.005955 -0.007018 -0.00833
Coordinates: (3/7)
* time (time) float64 2kB -5.12 -4.992 -4.864 ... 19.84 19.97 20.1
samples (time) int64 2kB 0 1 2 3 4 5 6 7 ... 191 192 193 194 195 196 197
... ...
* trial_type (trial_type) <U13 104B 'Tapping/Left' 'Tapping/Right'
Attributes:
data_type_group: processed HRF concentrationsAssert that the written and read HRFs are identical.
[18]:
assert (read_blockaverage.rename({"time" : "reltime"}) == rec["hrf_blockaverage"]).all()
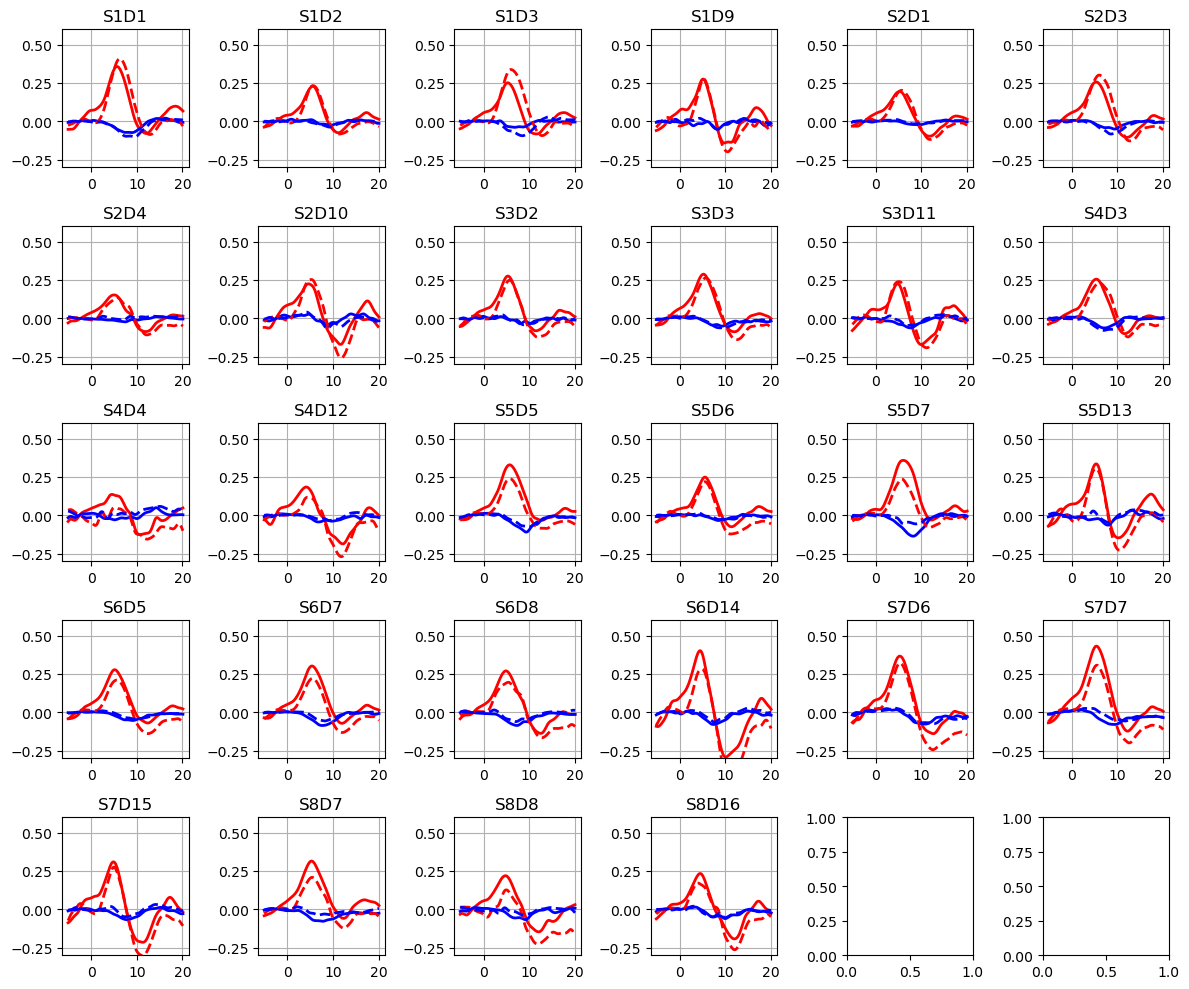
Plot the HRFs
[19]:
ba = read_blockaverage
f,ax = p.subplots(5,6, figsize=(12,10))
ax = ax.flatten()
for i_ch, ch in enumerate(ba.channel.values):
for ls, trial_type in zip(["-", "--"], ba.trial_type):
ax[i_ch].plot(ba.time, ba.sel(chromo="HbO", trial_type=trial_type, channel=ch), "r", lw=2, ls=ls)
ax[i_ch].plot(ba.time, ba.sel(chromo="HbR", trial_type=trial_type, channel=ch), "b", lw=2, ls=ls)
ax[i_ch].grid(1)
ax[i_ch].set_title(ch)
ax[i_ch].set_ylim(-.3, .6)
p.tight_layout()
Tidy up
[20]:
del tmpdir