Xarray Data Structures - an fNIRS example
This example illustrates the usage of xarray-based data structures for calculating the Beer-Lambert transformation.
[1]:
# This cells setups the environment when executed in Google Colab.
try:
import google.colab
!curl -s https://raw.githubusercontent.com/ibs-lab/cedalion/dev/scripts/colab_setup.py -o colab_setup.py
# Select branch with --branch "branch name" (default is "dev")
%run colab_setup.py
except ImportError:
pass
[2]:
import cedalion
import cedalion.nirs
import cedalion.xrutils
import cedalion.xrutils as xrutils
from cedalion.data import get_fingertapping_snirf_path
import numpy as np
import xarray as xr
import pint
import matplotlib.pyplot as p
import scipy.signal
import os.path
xr.set_options(display_max_rows=3, display_values_threshold=50)
np.set_printoptions(precision=4)
Loading raw CW-NIRS data from a SNIRF file
This notebook uses a finger-tapping dataset in BIDS layout provided by Rob Luke. It can can be downloaded via cedalion.data.
Load amplitude data from the snirf file.
[3]:
path_to_snirf_file = get_fingertapping_snirf_path()
recordings = cedalion.io.read_snirf(path_to_snirf_file)
rec = recordings[0] # there is only one NirsElement in this snirf file...
amp = rec["amp"] # ... which holds amplitude data
# restrict to first 60 seconds and fill in missing units
amp = amp.sel(time=amp.time < 60)
amp = amp.pint.dequantify().pint.quantify("V")
geo3d = rec.geo3d
[4]:
recordings
[4]:
[<Recording | timeseries: ['amp'], masks: [], stim: ['1.0', '15.0', '2.0', '3.0'], aux_ts: [], aux_obj: []>]
Amplitude data
[5]:
display(amp.round(4))
<xarray.DataArray (channel: 28, wavelength: 2, time: 469)> Size: 210kB
<Quantity([[[0.0914 0.091 0.091 ... 0.0903 0.0902 0.0899]
[0.1857 0.1864 0.1837 ... 0.1849 0.185 0.1847]]
[[0.2275 0.2297 0.2261 ... 0.2241 0.2243 0.2257]
[0.6355 0.6377 0.6298 ... 0.6223 0.6237 0.6272]]
[[0.1065 0.1066 0.1053 ... 0.1065 0.1062 0.1056]
[0.2755 0.2762 0.2727 ... 0.2737 0.2742 0.276 ]]
...
[[0.2028 0.1997 0.2005 ... 0.1998 0.2007 0.2026]
[0.4666 0.4554 0.4562 ... 0.4482 0.4511 0.4541]]
[[0.4885 0.4802 0.4818 ... 0.5005 0.5036 0.5045]
[0.8458 0.826 0.826 ... 0.8386 0.8441 0.8475]]
[[0.6305 0.6284 0.6287 ... 0.6373 0.638 0.6392]
[1.2286 1.2206 1.219 ... 1.2232 1.2259 1.2278]]], 'volt')>
Coordinates: (3/6)
* time (time) float64 4kB 0.0 0.128 0.256 0.384 ... 59.65 59.78 59.9
samples (time) int64 4kB 0 1 2 3 4 5 6 7 ... 462 463 464 465 466 467 468
... ...
* wavelength (wavelength) float64 16B 760.0 850.0
Attributes:
data_type_group: unprocessed rawMontage information
The geo3d DataArray maps labels to 3D positions, thus storing the location of optodes and landmarks.
[6]:
display_labels = ["S1", "S2", "D1", "D2", "NASION"] # for brevity show only these
geo3d.round(5).sel(label=display_labels)
[6]:
<xarray.DataArray (label: 5, pos: 3)> Size: 120B
<Quantity([[-0.0416 0.0268 0.1299]
[-0.0648 0.0581 0.0908]
[-0.0376 0.0632 0.1157]
[-0.0413 -0.0118 0.1349]
[ 0. 0.114 -0. ]], 'meter')>
Coordinates:
* label (label) <U6 120B 'S1' 'S2' 'D1' 'D2' 'NASION'
type (label) object 40B PointType.SOURCE ... PointType.LANDMARK
Dimensions without coordinates: posTo obtain channel distances, we can lookup amp’s source and detector coordinates in geo3d, subtract these and calculate the vector norm.
[7]:
dists = xrutils.norm(geo3d.loc[amp.source] - geo3d.loc[amp.detector], dim="pos")
display(dists.round(3))
<xarray.DataArray (channel: 28)> Size: 224B
<Quantity([0.039 0.039 0.041 0.008 0.037 0.038 0.037 0.007 0.04 0.037 0.008 0.041
0.034 0.008 0.039 0.039 0.041 0.008 0.037 0.037 0.037 0.008 0.04 0.037
0.007 0.041 0.033 0.008], 'meter')>
Coordinates:
* channel (channel) object 224B 'S1D1' 'S1D2' 'S1D3' ... 'S8D8' 'S8D16'
source (channel) object 224B 'S1' 'S1' 'S1' 'S1' ... 'S7' 'S8' 'S8' 'S8'
detector (channel) object 224B 'D1' 'D2' 'D3' 'D9' ... 'D7' 'D8' 'D16'Beer-Lambert transformation
Specify differential path length factors (DPF). Obtain a matrix of tabulated extinction coefficients for the wavelengths of our dataset and calculate the inverse. Cedalion offers dedicated functions for mBLL conversion ( nirs.int2od(), nirs.od2conc(), and nirs.beer-lambert() functions from the nirs subpackage) - but we do not use them here to better showcase how Xarrays work.
[8]:
dpf = xr.DataArray([6., 6.], dims="wavelength", coords={"wavelength" : [760., 850.]})
E = cedalion.nirs.get_extinction_coefficients("prahl", amp.wavelength)
Einv = cedalion.xrutils.pinv(E)
display(Einv.round(4))
<xarray.DataArray (wavelength: 2, chromo: 2)> Size: 32B <Quantity([[-0.0024 0.0037] [ 0.0055 -0.0021]], 'millimeter * molar')> Coordinates: * chromo (chromo) <U3 24B 'HbO' 'HbR' * wavelength (wavelength) float64 16B 760.0 850.0
[9]:
optical_density = -np.log( amp / amp.mean("time"))
conc = Einv @ (optical_density / ( dists * dpf))
display(conc.pint.to("micromolar").round(4))
<xarray.DataArray (chromo: 2, channel: 28, time: 469)> Size: 210kB
<Quantity([[[-0.0538 -0.1835 0.1612 ... -0.0725 -0.1064 -0.0939]
[-0.1971 -0.1772 -0.0504 ... 0.1388 0.0943 0.0257]
[ 0.0052 -0.0338 0.1269 ... 0.1558 0.0805 -0.1094]
...
[-0.4716 -0.084 -0.0781 ... 0.2843 0.1805 0.1309]
[-0.6011 -0.1653 -0.124 ... -0.0753 -0.1786 -0.2662]
[-1.1848 -0.5717 -0.392 ... -0.076 -0.2826 -0.3658]]
[[-0.168 -0.0692 -0.2042 ... -0.0249 0.0083 0.0364]
[-0.0429 -0.1653 -0.0254 ... 0.0097 0.0158 -0.0304]
[-0.0471 -0.0485 0.0284 ... -0.1066 -0.0428 0.0851]
...
[ 0.0045 0.0366 -0.0132 ... -0.1122 -0.1236 -0.2155]
[ 0.3211 0.3949 0.3327 ... -0.2165 -0.264 -0.2554]
[ 0.6814 0.6478 0.554 ... -0.4091 -0.3963 -0.487 ]]], 'micromolar')>
Coordinates: (3/6)
* chromo (chromo) <U3 24B 'HbO' 'HbR'
* time (time) float64 4kB 0.0 0.128 0.256 0.384 ... 59.65 59.78 59.9
... ...
detector (channel) object 224B 'D1' 'D2' 'D3' 'D9' ... 'D7' 'D8' 'D16'[10]:
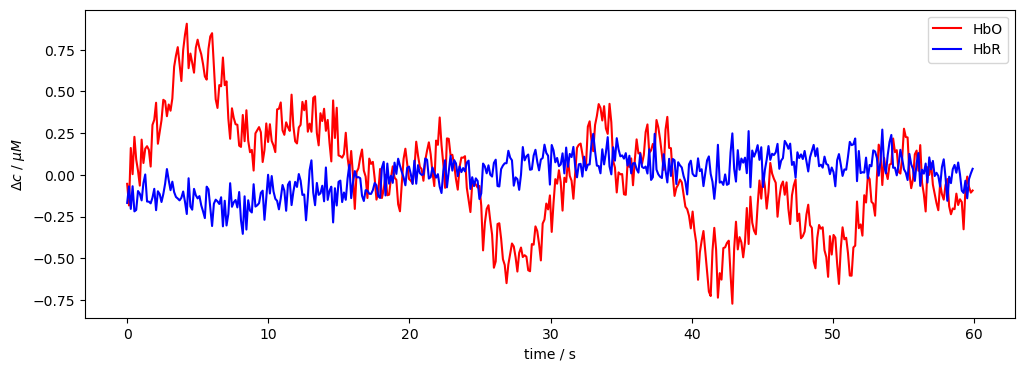
f,ax = p.subplots(1,1, figsize=(12,4))
ax.plot( conc.time, conc.sel(channel="S1D1", chromo="HbO").pint.to("micromolar"), "r-", label="HbO")
ax.plot( conc.time, conc.sel(channel="S1D1", chromo="HbR").pint.to("micromolar"), "b-", label="HbR")
p.legend()
ax.set_xlabel("time / s")
ax.set_ylabel("$\Delta c$ / $\mu M$");