GLM Basis Functions
[1]:
# This cells setups the environment when executed in Google Colab.
try:
import google.colab
!curl -s https://raw.githubusercontent.com/ibs-lab/cedalion/dev/scripts/colab_setup.py -o colab_setup.py
# Select branch with --branch "branch name" (default is "dev")
%run colab_setup.py
except ImportError:
pass
[2]:
import cedalion
import cedalion.models.glm.basis_functions as bf
import cedalion.models.glm.design_matrix as dm
import cedalion.dataclasses as cdc
import matplotlib.pyplot as p
import numpy as np
import xarray as xr
import matplotlib.pyplot as p
import cedalion.data
units = cedalion.units
xr.set_options(display_expand_data=False)
[2]:
<xarray.core.options.set_options at 0x7e420b30e410>
[3]:
# dummy time series
fs = 8.0
ts = cdc.build_timeseries(
np.random.random((100, 1, 2)),
dims=["time", "channel", "chromo"],
time=np.arange(100) / fs,
channel=["S1D1"],
value_units=units.uM,
time_units=units.s,
other_coords={'chromo' : ["HbO", "HbR"]}
)
display(ts)
<xarray.DataArray (time: 100, channel: 1, chromo: 2)> Size: 2kB
[µM] 0.3857 0.3946 0.647 0.2247 0.2743 ... 0.3566 0.0009899 0.7693 0.8115 0.2819
Coordinates:
* time (time) float64 800B 0.0 0.125 0.25 0.375 ... 12.0 12.12 12.25 12.38
samples (time) int64 800B 0 1 2 3 4 5 6 7 8 ... 91 92 93 94 95 96 97 98 99
* channel (channel) <U4 16B 'S1D1'
* chromo (chromo) <U3 24B 'HbO' 'HbR'[4]:
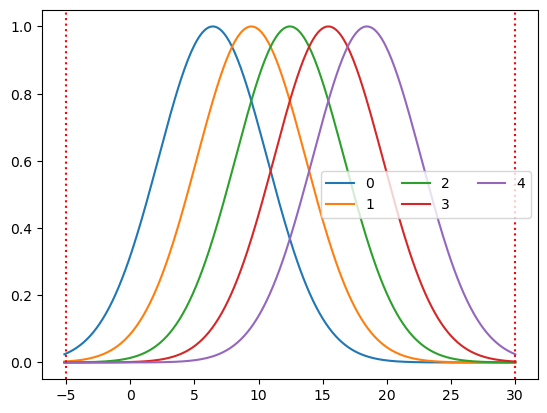
basis = bf.GaussianKernels(
t_pre=5 * units.s,
t_post=30 * units.s,
t_delta=3 * units.s,
t_std=3 * units.s,
)
hrf = basis(ts)
p.figure()
for i_comp, comp in enumerate(hrf.component.values):
p.plot(hrf.time, hrf[:, i_comp], label=comp)
p.axvline(-5, c="r", ls=":")
p.axvline(30, c="r", ls=":")
p.legend(ncols=3)
[4]:
<matplotlib.legend.Legend at 0x7e4209800790>
[5]:
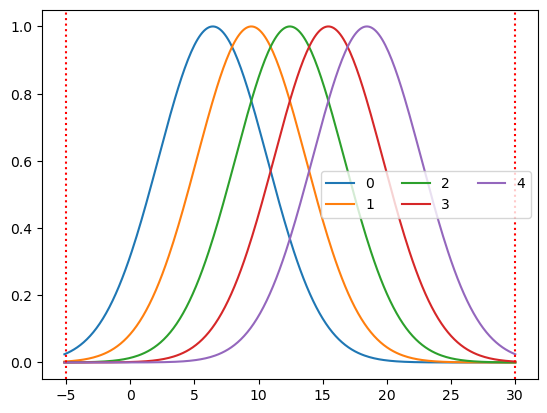
basis = bf.GaussianKernelsWithTails(
t_pre=5 * units.s,
t_post=30 * units.s,
t_delta=3 * units.s,
t_std=3 * units.s,
)
hrf = basis(ts)
p.figure()
for i_comp, comp in enumerate(hrf.component.values):
p.plot(hrf.time, hrf[:, i_comp], label=comp)
p.axvline(-5, c="r", ls=":")
p.axvline(30, c="r", ls=":")
p.legend(ncols=3)
[5]:
<matplotlib.legend.Legend at 0x7e42097ae610>
[6]:
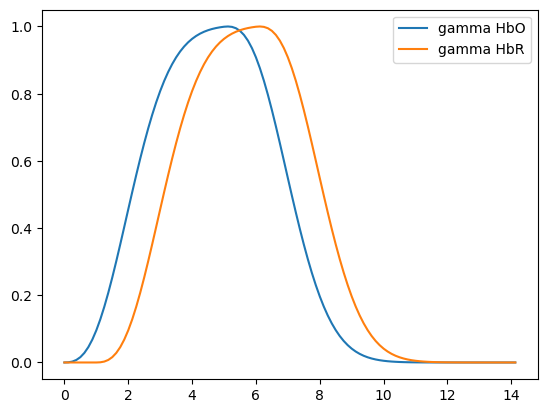
basis = bf.Gamma(
tau={"HbO": 0 * units.s, "HbR": 1 * units.s},
sigma=3 * units.s,
)
hrf = basis(ts)
display(hrf)
p.figure()
for i_comp, comp in enumerate(hrf.component.values):
for i_chromo, chromo in enumerate(hrf.chromo.values):
p.plot(hrf.time, hrf[:, i_comp, i_chromo], label=f"{comp} {chromo}")
p.legend()
<xarray.DataArray (time: 107, component: 1, chromo: 2)> Size: 2kB 0.0 0.0 0.004711 0.0 0.01875 ... 2.533e-07 3.574e-06 1.789e-07 2.601e-06 Coordinates: * time (time) float64 856B 0.0 0.125 0.25 0.375 ... 13.0 13.12 13.25 * chromo (chromo) <U3 24B 'HbO' 'HbR' * component (component) <U5 20B 'gamma'
[6]:
<matplotlib.legend.Legend at 0x7e4201593550>
[7]:
basis = bf.Gamma(
tau={"HbO": 0 * units.s, "HbR": 1 * units.s},
sigma=2 * units.s,
)
hrf = basis(ts)
display(hrf)
p.figure()
for i_comp, comp in enumerate(hrf.component.values):
for i_chromo, chromo in enumerate(hrf.chromo.values):
p.plot(hrf.time, hrf[:, i_comp, i_chromo], label=f"{comp} {chromo}")
p.legend()
<xarray.DataArray (time: 74, component: 1, chromo: 2)> Size: 1kB 0.0 0.0 0.01058 0.0 0.04181 ... 7.789e-06 8.836e-08 4.894e-06 5.155e-08 3.05e-06 Coordinates: * time (time) float64 592B 0.0 0.125 0.25 0.375 ... 8.75 8.875 9.0 9.125 * chromo (chromo) <U3 24B 'HbO' 'HbR' * component (component) <U5 20B 'gamma'
[7]:
<matplotlib.legend.Legend at 0x7e420042c5d0>
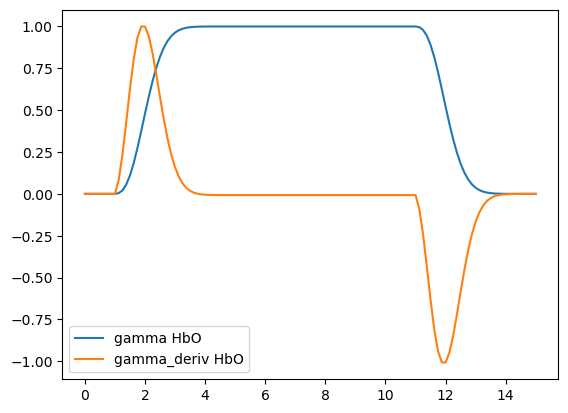
[8]:
basis = bf.GammaDeriv(
tau=2 * units.s,
sigma=2 * units.s,
)
hrf = basis(ts)
display(hrf)
p.figure()
for i_comp, comp in enumerate(hrf.component.values):
for i_chromo, chromo in enumerate(["HbO"]):
p.plot(hrf.time, hrf[:, i_comp, i_chromo], label=f"{comp} {chromo}")
p.legend()
<xarray.DataArray (time: 82, component: 2, chromo: 2)> Size: 3kB 0.0 0.0 0.0 0.0 0.0 0.0 ... -2.311e-05 3.05e-06 3.05e-06 -1.466e-05 -1.466e-05 Coordinates: * time (time) float64 656B 0.0 0.125 0.25 0.375 ... 9.875 10.0 10.12 * chromo (chromo) <U3 24B 'HbO' 'HbR' * component (component) <U11 88B 'gamma' 'gamma_deriv'
[8]:
<matplotlib.legend.Legend at 0x7e42004ce610>
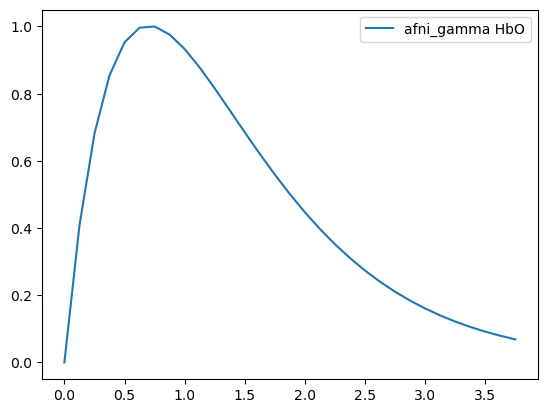
[9]:
basis = bf.AFNIGamma(
p=1,
q=0.7 * units.s,
)
hrf = basis(ts)
display(hrf)
p.figure()
for i_comp, comp in enumerate(hrf.component.values):
for i_chromo, chromo in enumerate(["HbO"]):
p.plot(hrf.time, hrf[:, i_comp, i_chromo], label=f"{comp} {chromo}")
p.legend()
<xarray.DataArray (time: 31, component: 1, chromo: 2)> Size: 496B 0.0 0.0 0.407 0.407 0.6809 0.6809 ... 0.0918 0.07953 0.07953 0.06882 0.06882 Coordinates: * time (time) float64 248B 0.0 0.125 0.25 0.375 ... 3.375 3.5 3.625 3.75 * chromo (chromo) <U3 24B 'HbO' 'HbR' * component (component) <U10 40B 'afni_gamma'
[9]:
<matplotlib.legend.Legend at 0x7e4200346b50>
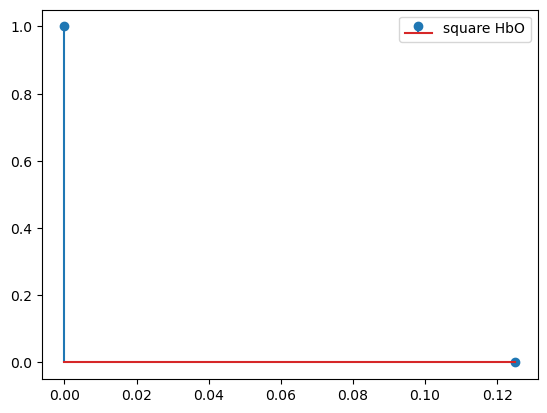
[10]:
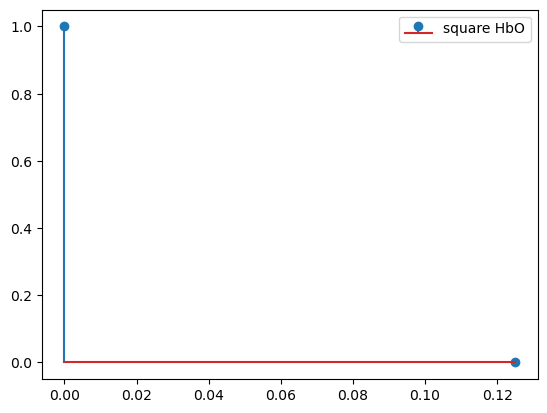
basis = bf.DiracDelta()
hrf = basis(ts)
display(hrf)
p.figure()
for i_comp, comp in enumerate(hrf.component.values):
for i_chromo, chromo in enumerate(["HbO"]):
p.stem(hrf.time, hrf[:, i_comp, i_chromo], label=f"{comp} {chromo}")
p.legend()
<xarray.DataArray (time: 2, component: 1, chromo: 2)> Size: 32B 1.0 1.0 0.0 0.0 Coordinates: * time (time) float64 16B 0.0 0.125 * chromo (chromo) <U3 24B 'HbO' 'HbR' * component (component) <U6 24B 'square'
[10]:
<matplotlib.legend.Legend at 0x7e42004a7490>